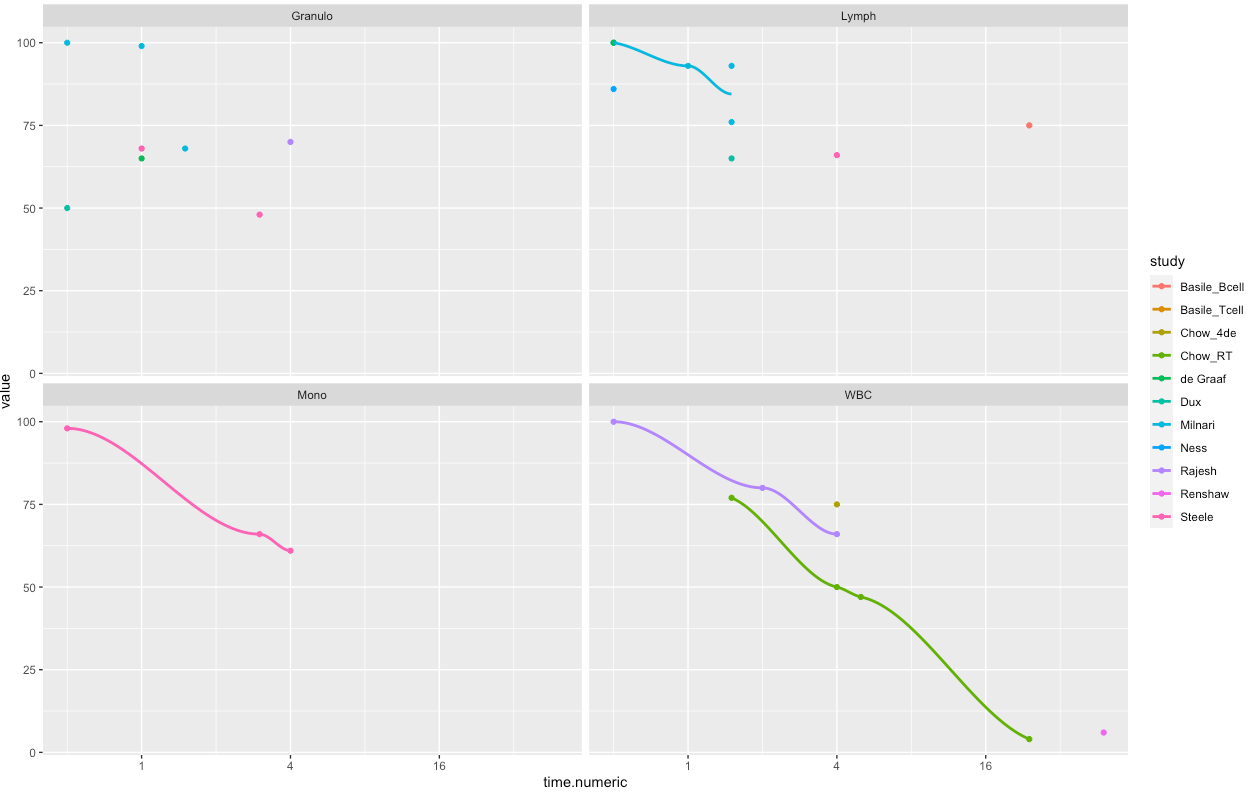
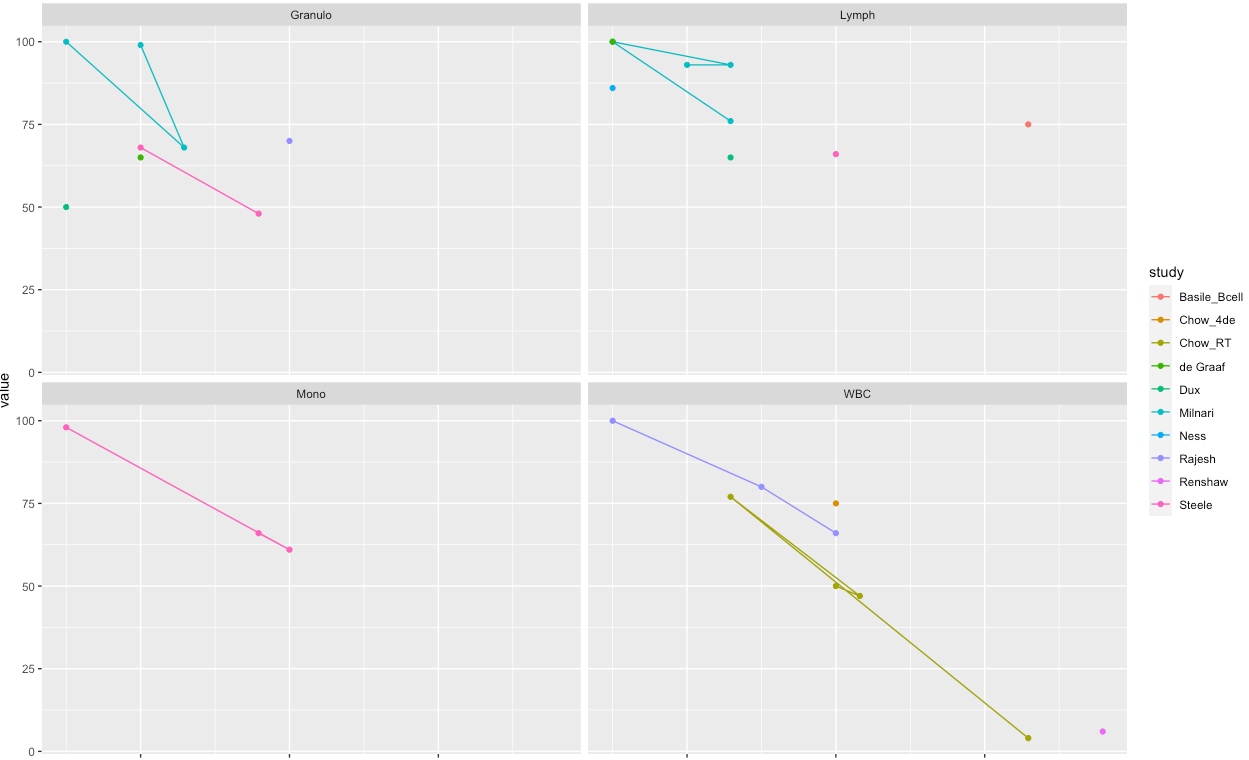
Let's say I have
'data.frame': 75 obs. of 4 variables:
$ study : chr "Dux" "Steele" "Ness" "Dux" ...
$ celltype : chr "Lymph" "Granulo" "Lymph" "Lymph" ...
$ value : int NA NA 86 NA NA NA NA NA 76 100 ...
$ time.numeric: num 4 5 0.5 6 4 2 3 24 1.5 0.5 ...
I aim to connect all geom_point() by a geom_smooth(), also when breaks due to NA are present. Why is geom_smooth() not connecting all geom_points()? It seems that it only work for some of the facet_wraps despite multiple geom_points() are present in all facets
a <– structure(list(study = c("Milnari", "Rajesh", "Steele", "de Graaf",
"Chow_RT", "Steele", "Milnari", "Dux", "de Graaf", "Basile_Bcell",
"Steele", "de Graaf", "Renshaw", "Renshaw", "Milnari", "Rajesh",
"Steele", "Rajesh", "Rajesh", "Dux", "Ness", "Basile_Tcell",
"Rajesh", "de Graaf", "Milnari", "Dux", "Ness", "de Graaf", "de Graaf",
"Rajesh", "Rajesh", "Chow_RT", "Steele", "de Graaf", "Milnari",
"Ness", "Dux", "Dux", "Ness", "Basile_Bcell", "Dux", "Milnari",
"Renshaw", "Basile_Bcell", "Steele", "Renshaw", "Rajesh", "Rajesh",
"Milnari", "de Graaf"), celltype = c("Lymph", "Granulo", "Granulo",
"Mono", "WBC", "Mono", "Lymph", "Lymph", "Lymph", "Lymph", "Granulo",
"Granulo", "WBC", "WBC", "Lymph", "WBC", "Lymph", "Granulo",
"Granulo", "Granulo", "Lymph", "Lymph", "Granulo", "Mono", "Lymph",
"Granulo", "Lymph", "Mono", "Granulo", "WBC", "WBC", "WBC", "Granulo",
"Lymph", "Granulo", "Lymph", "Lymph", "Lymph", "Lymph", "Lymph",
"Lymph", "Lymph", "WBC", "Lymph", "Granulo", "WBC", "WBC", "Granulo",
"Granulo", "Granulo"), value = c(93L, 81L, 50L, NA, NA, NA, NA,
88L, NA, NA, 42L, NA, 6L, NA, 93L, NA, 69L, 100L, NA, 25L, NA,
85L, 70L, NA, NA, 10L, 80L, NA, NA, NA, 80L, 85L, NA, NA, NA,
NA, NA, NA, NA, 75L, NA, NA, NA, NA, 48L, 30L, NA, NA, NA, 58L
), time.numeric = c(1.5, 2, 2, 4, 48, 5, 2, 0.5, 4, 2, 4, 4,
48, 4, 1, 6, 3, 0.5, 1, 1, 24, 24, 4, 6, 48, 1.5, 1, 48, 6, 3,
2, 1, 1.5, 2, 5, 2, 2, 5, 6, 24, 48, 2, 2, 4, 3, 24, 24, 3, 6,
5)), class = "data.frame", row.names = c(NA, -50L))
My attempt:
ggplot(a, aes(x = time.numeric, y = value,
color = study, fill = study))
geom_point()
scale_x_continuous(trans = "log2")
facet_wrap(~ celltype)
geom_smooth(se = FALSE)
I seem to receive a warning(), however I cannot relate it to the issue.
> warnings()
Warning messages:
1: Removed 46 rows containing non-finite values (stat_smooth).
2: In simpleLoess(y, x, w, span, degree = degree, parametric = parametric, ... :
span too small. fewer data values than degrees of freedom.
3: In simpleLoess(y, x, w, span, degree = degree, parametric = parametric, ... :
pseudoinverse used at -1.0079
CodePudding user response:
As suggested by Jon Spring, the solution was geom_path() with prior removal of NA
ggplot(filter(a, !is.na(value)), aes(x = time.numeric, y = value,
color = study, fill = study))
geom_path()
geom_point()
scale_x_continuous(trans = "log2")
facet_wrap(~ celltype)