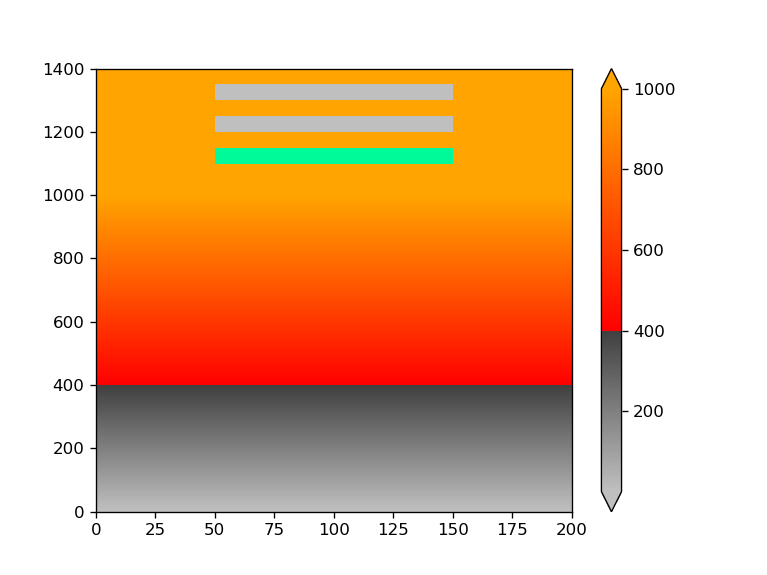
I'd like to use a custom (blue) color on a plot where the data is zero. I have tried the set_under method, but failed. The desired output would be a blue line at the bottom and a two blue square at the upper part of the graph. Any help is appreciated.
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.colors import LinearSegmentedColormap,TwoSlopeNorm
# TwoSlopeNorm see:
# https://matplotlib.org/devdocs/tutorials/colors/colormapnorms.html#sphx-glr-tutorials-colors-colormapnorms-py
red2orange = np.array([np.linspace(1, 1, 256),
np.linspace(0, 165/256, 256),
np.linspace(0, 0, 256),
np.ones(256)]).T
grey2black = np.array([np.linspace(0.75, 0.25, 256),
np.linspace(0.75, 0.25, 256),
np.linspace(0.75, 0.25, 256),
np.ones(256)]).T
all_colors = np.vstack((grey2black,red2orange))
cmap = LinearSegmentedColormap.from_list('two_slope_cmap', all_colors)
divnorm = TwoSlopeNorm(vmin=1, vcenter=400, vmax=1000)
# seting bad and under
cmap.set_bad('mediumspringgreen')
cmap.set_under('blue')
#fake data
data = np.arange(1400)[:,None]* np.ones(200)
data[ 1100:1150, 50:150] = np.nan # bad data
data[ 1200:1250, 50:150] = 0 # zero data
data[ 1300:1350, 50:150] = -1 # under data
# plot
f,a = plt.subplots()
raster = a.pcolormesh(data,cmap=cmap, norm=divnorm)
cbar = f.colorbar(raster,ax=a, extend='both')
CodePudding user response:
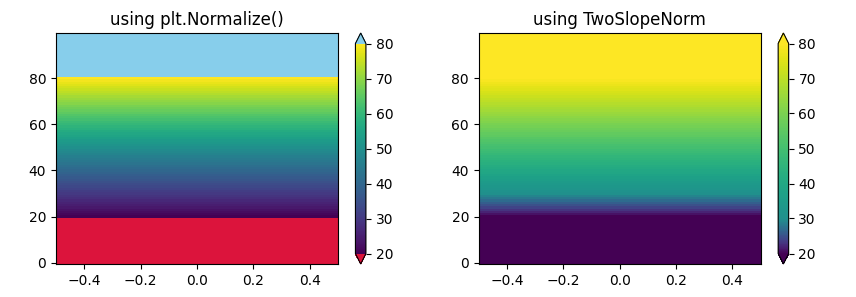
For some unknown reason, TwoSlopeNorm doesn't seem to honor the under nor over colors. Changing the code to use plt.Normalize() instead of TwoSlopeNorm() indicates that for that norm, the under color works as expected.
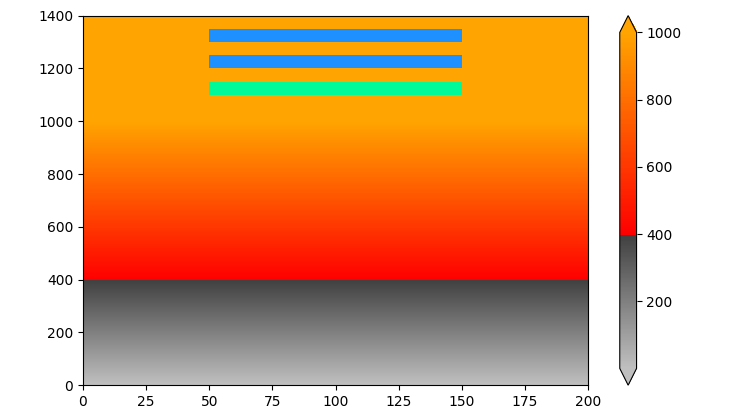
A workaround is to draw the pcolormesh a second time, only for the under color. A drawback is that the under color isn't shown in the colorbar extension.
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.colors import LinearSegmentedColormap, ListedColormap, TwoSlopeNorm
red2orange = np.array([np.linspace(1, 1, 256),
np.linspace(0, 165 / 256, 256),
np.linspace(0, 0, 256),
np.ones(256)]).T
grey2black = np.array([np.linspace(0.75, 0.25, 256),
np.linspace(0.75, 0.25, 256),
np.linspace(0.75, 0.25, 256),
np.ones(256)]).T
all_colors = np.vstack((grey2black, red2orange))
cmap = LinearSegmentedColormap.from_list('two_slope_cmap', all_colors)
divnorm = TwoSlopeNorm(vmin=1, vcenter=400, vmax=1000)
# seting bad and under
cmap.set_bad('mediumspringgreen')
cmap.set_under('dodgerblue') # this doesn't seem to be used with a TwoSlopeNorm
# fake data
data = np.arange(1400)[:, None] * np.ones(200)
data[1100:1150, 50:150] = np.nan # bad data
data[1200:1250, 50:150] = 0 # zero data
data[1300:1350, 50:150] = -1 # under data
# plot
f, a = plt.subplots()
raster = a.pcolormesh(data, cmap=cmap, norm=divnorm)
cbar = f.colorbar(raster, ax=a, extend='both')
# draw the mesh a second time, only for the under color
a.pcolormesh(np.where(data < 1, 0, np.nan), cmap=ListedColormap([cmap.get_under()]))
plt.show()
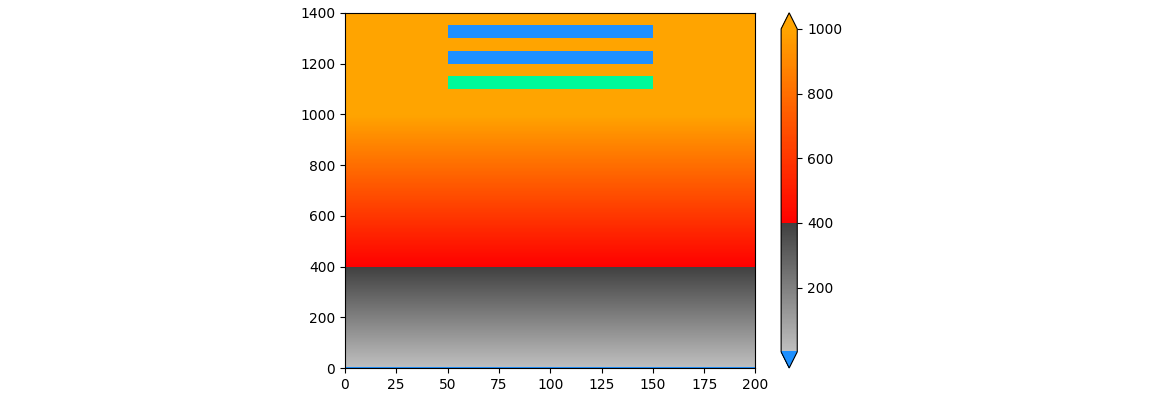
Another workaround is to change the colormap itself, setting the lowest color to the desired under color. A drawback is that vmin needs to move a bit. (The distance seems to be 1/127 of the distance between vmin and vcenter. Internally, the colormap maintains 256 colors, with the color for vcenter at position 128.)
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.colors import LinearSegmentedColormap, TwoSlopeNorm, to_rgba
red2orange = np.array([np.linspace(1, 1, 256),
np.linspace(0, 165 / 256, 256),
np.linspace(0, 0, 256),
np.ones(256)]).T
grey2black = np.array([np.linspace(0.75, 0.25, 256),
np.linspace(0.75, 0.25, 256),
np.linspace(0.75, 0.25, 256),
np.ones(256)]).T
all_colors = np.vstack((grey2black, red2orange))
all_colors[0, :] = to_rgba('dodgerblue')
cmap = LinearSegmentedColormap.from_list('two_slope_cmap', all_colors)
vmin = 1
vcenter = 400
vmax = 1000
divnorm = TwoSlopeNorm(vmin=vmin-(vcenter-vmin)/127, vcenter=vcenter, vmax=vmax)
# seting bad and under
cmap.set_bad('mediumspringgreen')
cmap.set_under('red') # this doesn't seem to be used with a TwoSlopeNorm
# fake data
data = np.arange(1400)[:, None] * np.ones(200)
data[1100:1150, 50:150] = np.nan # bad data
data[1200:1250, 50:150] = 0 # zero data
data[1300:1350, 50:150] = -1 # under data
# plot
f, a = plt.subplots()
raster = a.pcolormesh(data, cmap=cmap, norm=divnorm)
cbar = f.colorbar(raster, ax=a, extend='both')
plt.show()
PS: The following code tries to visualize how the two norms treat the under color differently:
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.colors import TwoSlopeNorm
fig, (ax1, ax2) = plt.subplots(ncols=2, figsize=(10, 3))
data = np.arange(0, 100)[:, None]
cmap = plt.get_cmap('viridis').copy()
cmap.set_under('crimson')
cmap.set_over('skyblue')
norm1 = plt.Normalize(20, 80)
im1 = ax1.imshow(data, cmap=cmap, norm=norm1, aspect='auto', origin='lower', interpolation='nearest')
plt.colorbar(im1, ax=ax1, extend='both')
ax1.set_title('using plt.Normalize()')
norm2 = TwoSlopeNorm(vmin=20, vcenter=30, vmax=80)
im2 = ax2.imshow(data, cmap=cmap, norm=norm2, aspect='auto', origin='lower', interpolation='nearest')
plt.colorbar(im2, ax=ax2, extend='both')
ax2.set_title('using TwoSlopeNorm')
plt.show()
PPS: Looking into the source of TwoSlopeNorm at github, the problem seems to be solved for the next version of matplotlib (current version is 3.4.3). So, you could try to install the development version. (The change involves adding left=-np.inf, right=np.inf as parameter to np.interp in the __call__ method of the TwoSlopeNorm class in colors.py.