The 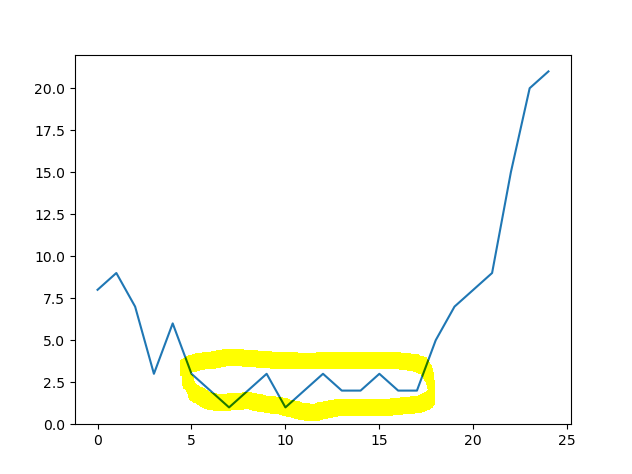
How can I identify the start and end index values of the less noisy and lower valued period marked in yellow?
Here is the test data:
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
arr = np.array([8,9,7,3,6,3,2,1,2,3,1,2,3,2,2,3,2,2,5,7,8,9,15,20,21])
plt.plot(arr)
plt.show()
CodePudding user response:
Well if you just want that 'area', you need some way of finding points within certain bounds. How can we do that? Well, we should probably start by finding the minimum of the array and then finding other values in that same array that fall within the specified deviation:
def lows(arr, dev=0):
lim = min(arr) dev
pts = []
for i,e in enumerate(arr):
if e <= lim:
pts.append((i,e))
return pts
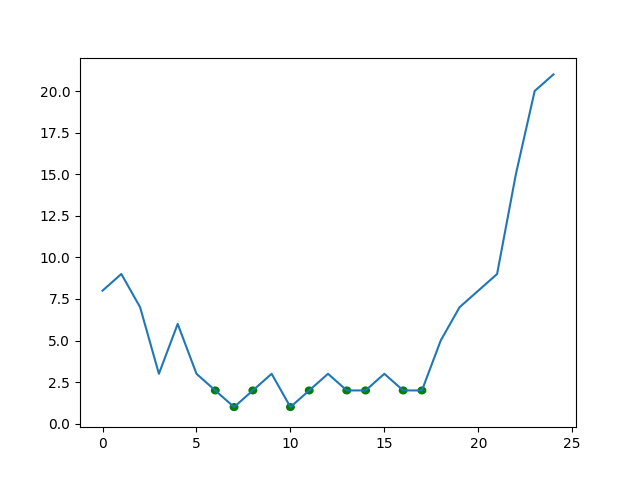
The above function returns a list of points that fall within the specified bounds. The lower bound is the minimum of the input array and the upper bound is the minimum value plus the deviation you will supply. For example, if you want all points within 1 of the lowest value:
plt.plot(arr)
for pt in lows(arr, 1):
circle = plt.Circle(pt, 0.2, color='g')
plt.gca().add_patch(circle)
plt.show()
CodePudding user response:
For a given point, we can decide to keep/mask it based on certain criteria:
- Are its neighbors are within some delta?
- Is it within some threshold of the minimum?
- Is it in a contiguous block?
Note: Since you tagged and imported pandas, I'll use pandas for convenience, but the same ideas can be implemented with pure numpy/matplotlib.
If all lower periods are around the same level
Then a simple approach is to use a neighbor delta with minimum threshold (though be careful of outliers in the real data):
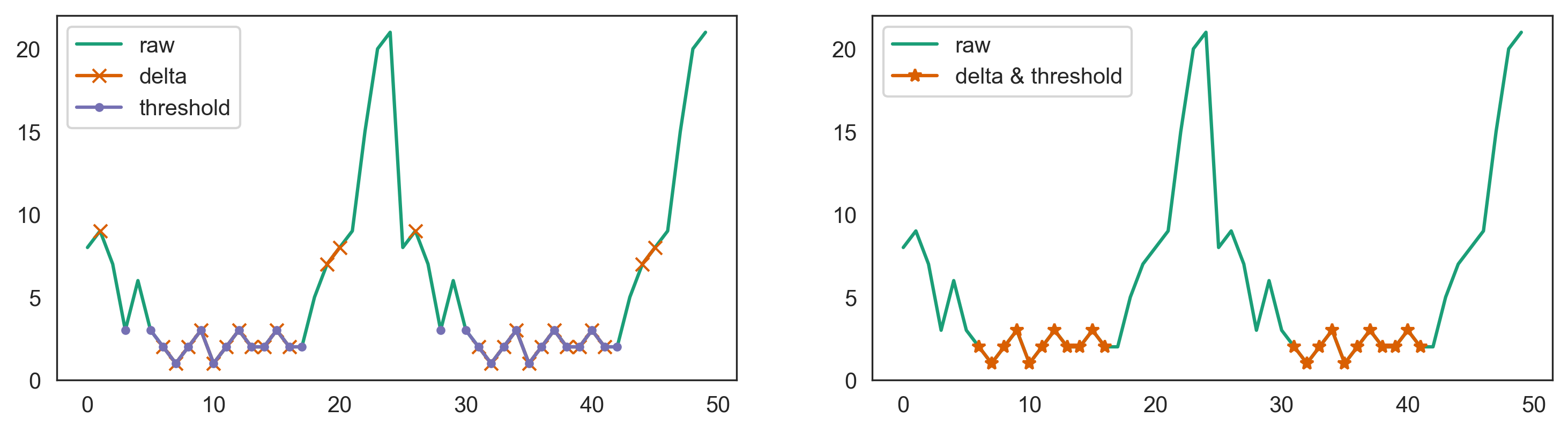
s = pd.Series(np.hstack([arr, arr]))
delta = 2
threshold = s.std()
mask_delta = s.diff().abs().le(delta) & s.diff(-1).abs().le(delta)
mask_threshold = s < s.min() threshold
s.plot(label='raw')
s.where(mask_threshold & mask_delta).plot(marker='*', label='delta & threshold')
If the lower periods are at different levels
Then a global minimum threshold won't work since some periods will be too high. In this case try a neighbor delta with contiguous blocks:
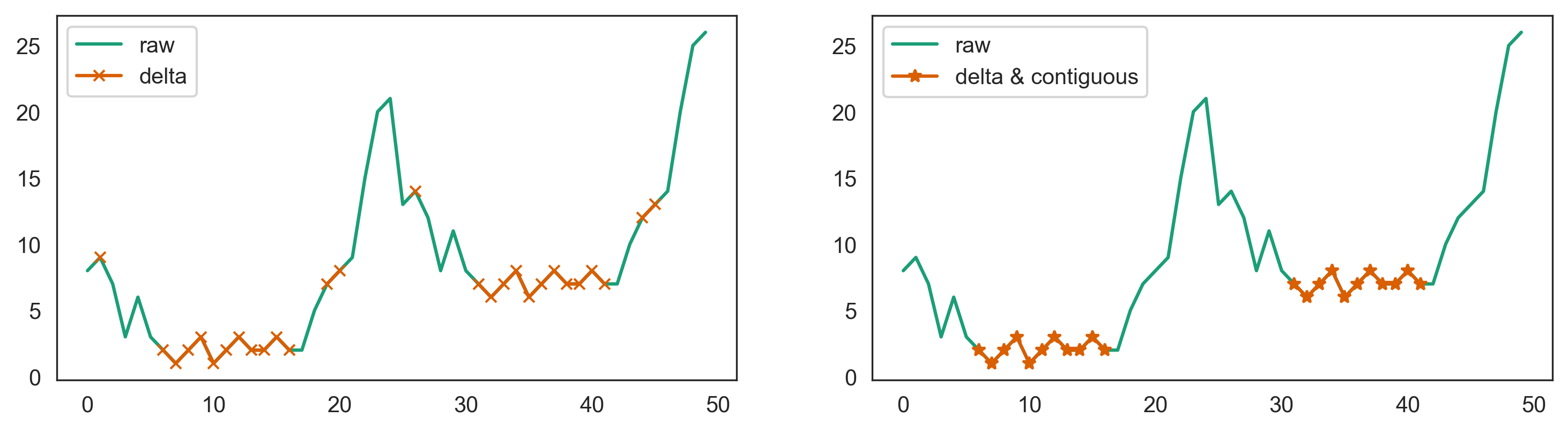
s = pd.Series(np.hstack([arr, arr 5]))
delta = 2
blocksize = 10
mask_delta = s.diff().abs().le(delta) & s.diff(-1).abs().le(delta)
masked = s.where(mask_delta)
groups = masked.isnull().cumsum()
blocksizes = masked.groupby(groups).transform('count').mask(masked.isnull())
mask_contiguous = blocksizes >= blocksize
s.plot(label='raw')
s.where(mask_contiguous).plot(marker='*', label='delta & contiguous')
