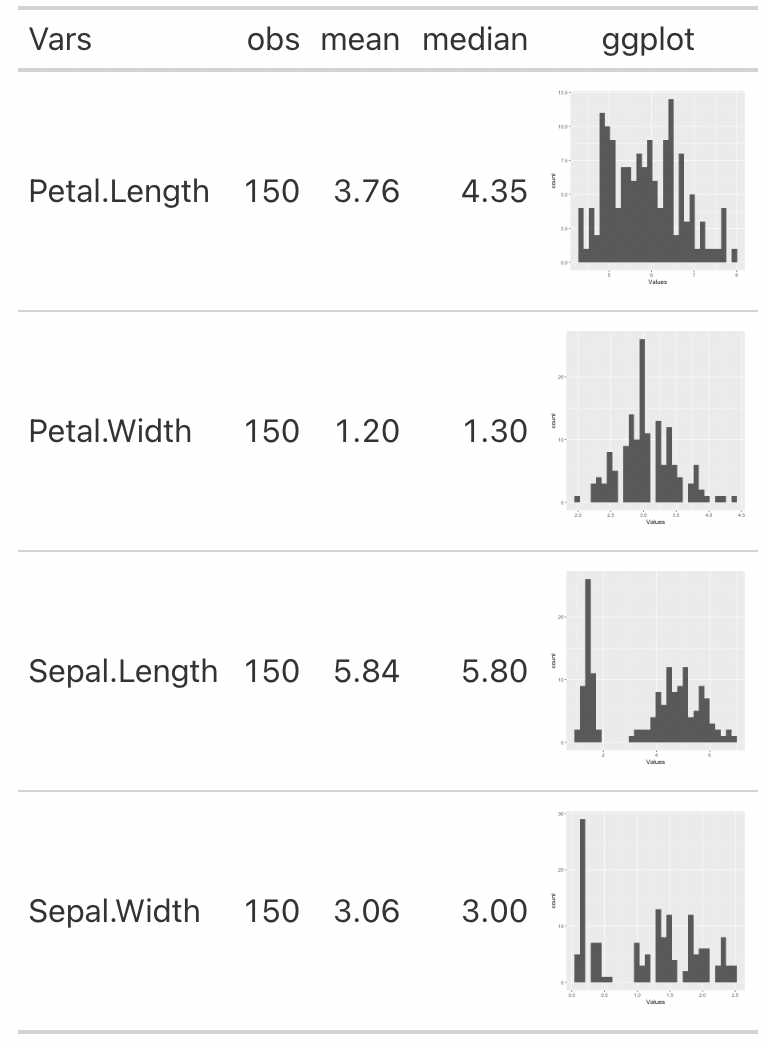
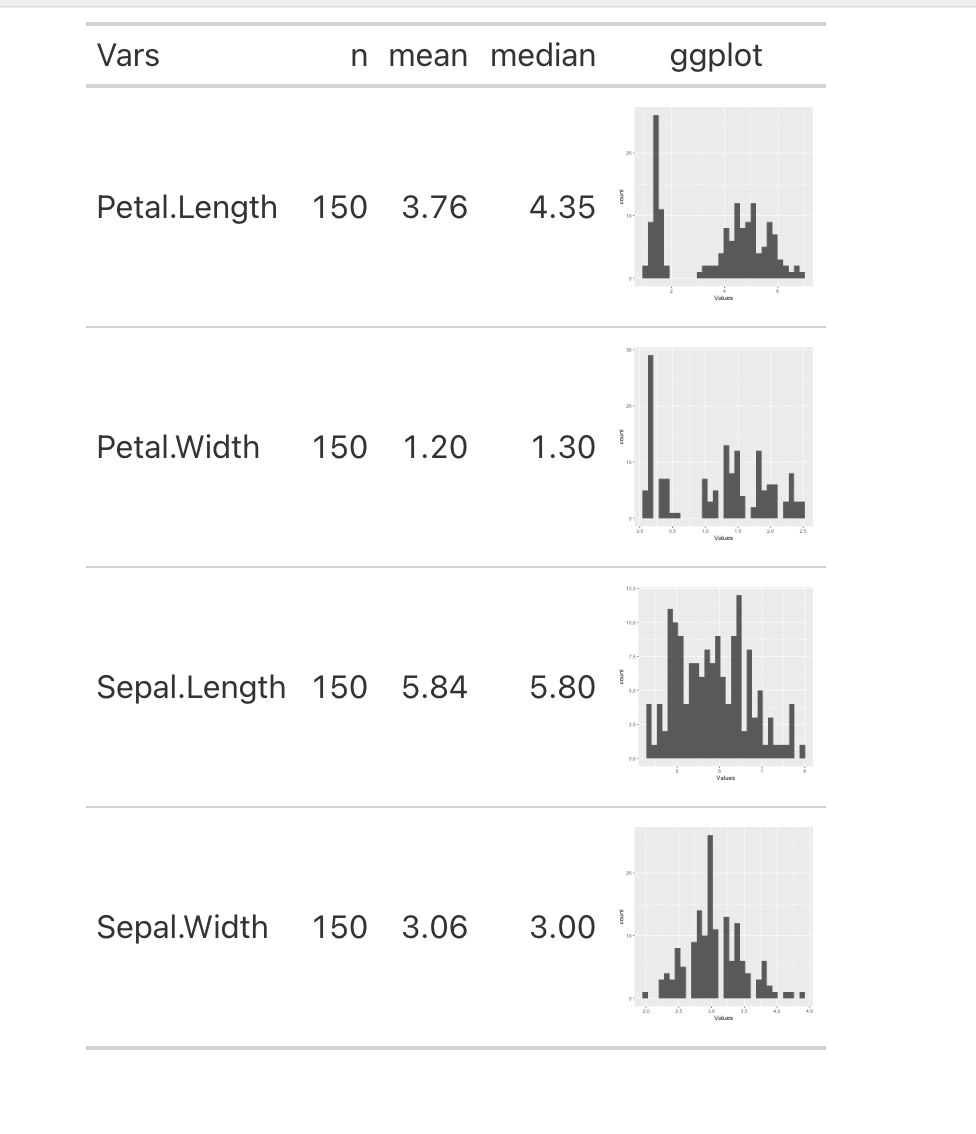
I want to create a gt table where I see some metrics like number of observations, mean and median, and I want a column with its histogram. For this question I will use the iris dataset.
I have recently learned how to put a plot in a tibble using this code:
library(dplyr)
library(tidyr)
library(purrr)
library(gt)
my_tibble <- iris %>%
pivot_longer(-Species,
names_to = "Vars",
values_to = "Values") %>%
group_by(Vars) %>%
summarise(obs = n(),
mean = round(mean(Values),2),
median = round(median(Values),2),
plots = list(ggplot(cur_data(), aes(Values)) geom_histogram()))
Now I want to use the plots column for plotting an histogram per variable, so I have tried this:
my_tibble %>%
mutate(ggplot = NA) %>%
gt() %>%
text_transform(
locations = cells_body(vars(ggplot)),
fn = function(x) {
map(.$plots,ggplot_image)
}
)
But it returns me an error:
Error in body[[col]][stub_df$rownum_i %in% loc$rows] <- fn(body[[col]][stub_df$rownum_i %in% :
replacement has length zero
The gt table should be like this:
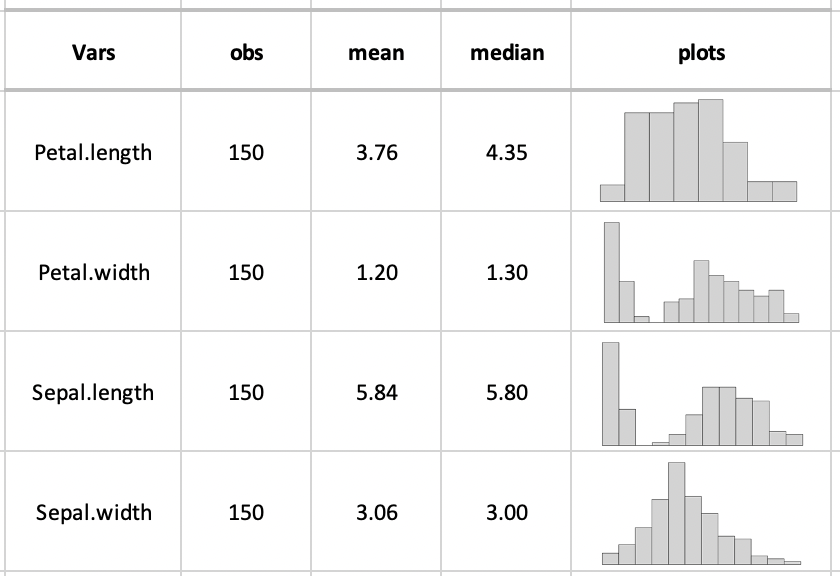
Any help will be greatly appreciated.
CodePudding user response:
Update: See comments:
For your purposes in accordance with a shiny app you may use summarytools see here: 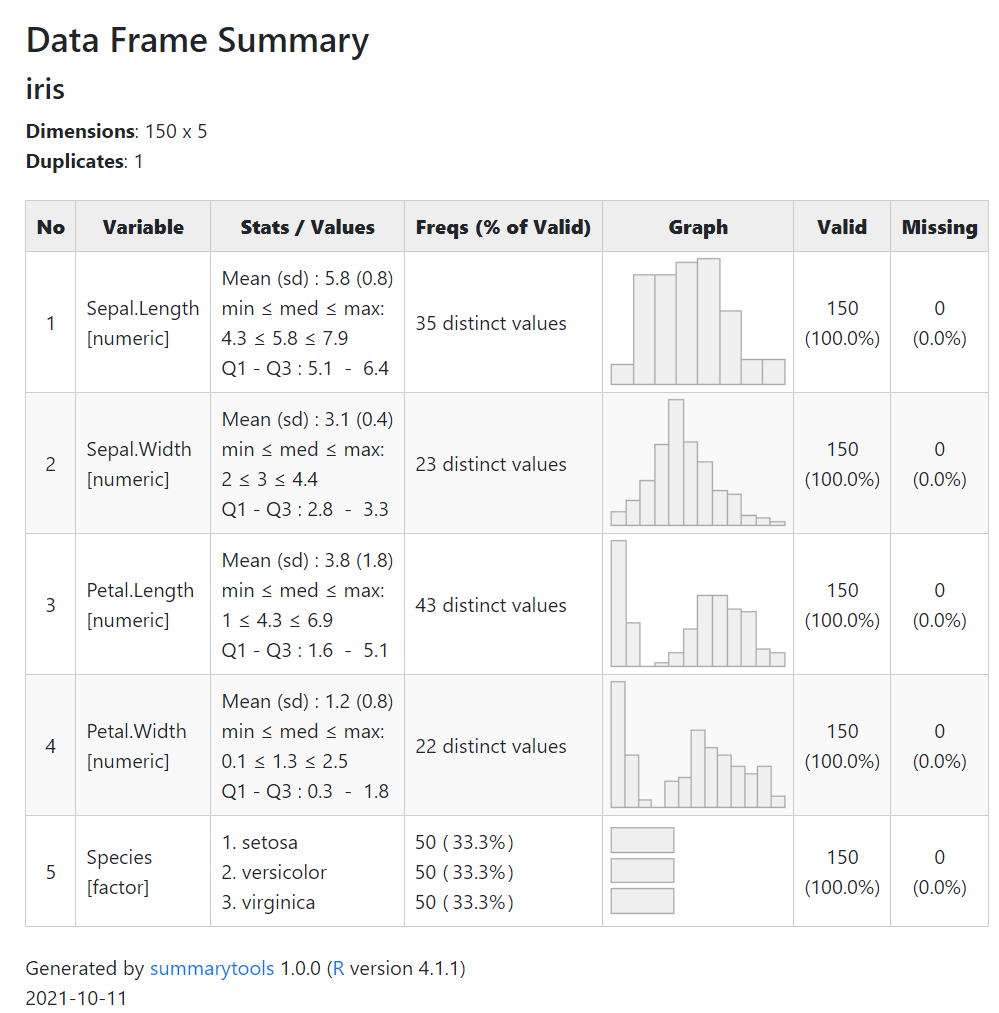
Try this:
library(skimr)
skim(iris)
skim_variable n_missing complete_rate mean sd p0 p25 p50 p75 p100 hist
* <chr> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr>
1 Sepal.Length 0 1 5.84 0.828 4.3 5.1 5.8 6.4 7.9 ▆▇▇▅▂
2 Sepal.Width 0 1 3.06 0.436 2 2.8 3 3.3 4.4 ▁▆▇▂▁
3 Petal.Length 0 1 3.76 1.77 1 1.6 4.35 5.1 6.9 ▇▁▆▇▂
4 Petal.Width 0 1 1.20 0.762 0.1 0.3 1.3 1.8 2.5 ▇▁▇▅▃
CodePudding user response:
After reviewing the excellent ideas from @akrun and @TarJae, I have this solution that gives the required gt table:
plots <- iris %>%
pivot_longer(-Species,
names_to = "Vars",
values_to = "Values") %>%
group_by(Vars) %>%
nest() %>%
mutate(plot = map(data,
function(df) df %>%
ggplot(aes(Values))
geom_histogram())) %>%
select(-data)
iris %>%
pivot_longer(-Species,
names_to = "Vars",
values_to = "Values") %>%
group_by(Vars) %>%
summarise(obs = n(),
mean = round(mean(Values),2),
median = round(median(Values),2)) %>%
mutate(ggplot = NA) %>%
gt() %>%
text_transform(
locations = cells_body(vars(ggplot)),
fn = function(x) {
map(plots$plot, ggplot_image, height = px(100))
}
)
I had to create the plot outside the output table, so I could call it in the gt table.
CodePudding user response:
We need to loop over the plots
library(dplyr)
library(tidyr)
library(purrr)
library(gt)
library(ggplot2)
iris %>%
pivot_longer(-Species,
names_to = "Vars",
values_to = "Values") %>%
nest_by(Vars) %>%
mutate(n = nrow(data),
mean = round(mean(data$Values), 2),
median = round(median(data$Values), 2),
plots = list(ggplot(data, aes(Values)) geom_histogram()), .keep = "unused") %>%
ungroup %>%
mutate(ggplot = NA) %>%
{dat <- .
dat %>%
select(-plots) %>%
gt() %>%
text_transform(locations = cells_body(c(ggplot)),
fn = function(x) {
map(dat$plots, ggplot_image, height = px(100))
}
)
}