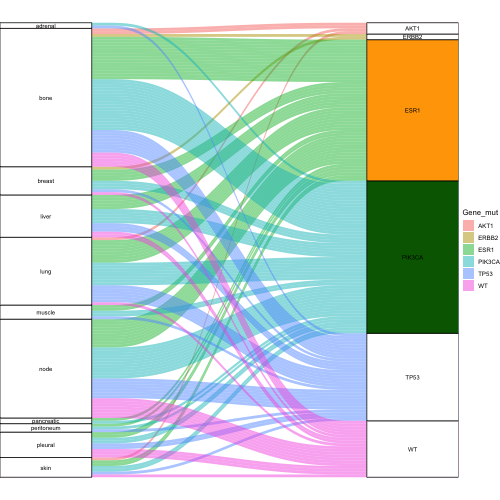
And I would like that the "strings" were the same color as the 2nd column, i.e. for ESR1 I would like the orange string, and for PIK3CA green.
Any idea about how can I manage with scale_fill_manual or any other argument?
Thanks!
My code:
colorfill <- c("white", "white", "darkgreen", "orange", "white", "white", "white", "white", "white", "white", "white", "white", "white", "white", "white", "white", "white")
ggplot(data = Allu,
aes(axis1 = Gene_mut, axis2 = Metastasis_Location, y = Freq))
geom_alluvium(aes(fill = Gene_mut),
curve_type = "quintic")
geom_stratum(width = 1/4, fill = colorfill)
geom_text(stat = "stratum", size = 3,
aes(label = after_stat(stratum)))
scale_x_discrete(limits = c("Metastasis_Location", "Gene_mut"),
expand = c(0.05, .05))
theme_void()
My data:
structure(list(Metastasis_Location = structure(c(1L, 1L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 4L, 4L, 4L, 4L, 4L, 4L,
4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 5L, 5L, 5L, 5L, 5L, 5L, 5L,
5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L,
5L, 6L, 6L, 6L, 6L, 6L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L,
7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L,
7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 8L, 8L, 9L, 9L, 9L, 10L,
10L, 10L, 10L, 10L, 10L, 10L, 10L, 10L, 11L, 11L, 11L, 11L, 11L,
11L, 11L), .Label = c("adrenal", "bone", "breast", "liver", "lung",
"muscle", "node", "pancreatic", "peritoneum", "pleural", "skin"
), class = "factor"), T0_T2_THERAPY_COD = structure(c(2L, 2L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 1L, 1L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L,
1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L), .Label = c("A",
"F"), class = "factor"), T0_T2_PD_event = structure(c(2L, 2L,
1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L,
2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 1L, 1L, 2L, 2L, 2L, 2L, 2L), .Label = c("No Progression",
"Progression"), class = "factor"), Gene_mut = structure(c(4L,
5L, 1L, 3L, 4L, 1L, 2L, 3L, 3L, 3L, 3L, 3L, 4L, 4L, 4L, 4L, 4L,
4L, 4L, 5L, 5L, 5L, 6L, 3L, 6L, 6L, 6L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 5L, 5L, 5L, 5L,
5L, 6L, 2L, 3L, 4L, 4L, 3L, 3L, 3L, 4L, 5L, 6L, 3L, 6L, 3L, 3L,
3L, 3L, 4L, 4L, 4L, 4L, 4L, 5L, 5L, 5L, 6L, 3L, 4L, 4L, 5L, 6L,
1L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 4L, 4L, 4L, 4L, 4L, 4L, 5L, 5L,
5L, 5L, 5L, 3L, 4L, 3L, 4L, 5L, 6L, 3L, 3L, 4L, 5L, 6L, 6L, 6L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L,
4L, 4L, 5L, 5L, 5L, 5L, 5L, 5L, 6L, 6L, 6L, 3L, 4L, 3L, 4L, 5L,
6L, 3L, 4L, 5L, 6L, 3L, 4L, 5L, 6L, 1L, 6L, 3L, 3L, 4L, 4L, 5L
), .Label = c("AKT1", "ERBB2", "ESR1", "PIK3CA", "TP53", "WT"
), class = "factor"), LABO_ID = structure(c(45L, 8L, 13L, 11L,
11L, 26L, 7L, 15L, 23L, 26L, 35L, 39L, 7L, 19L, 26L, 32L, 33L,
35L, 39L, 15L, 19L, 35L, 1L, 37L, 34L, 43L, 47L, 3L, 10L, 18L,
20L, 28L, 31L, 36L, 42L, 9L, 10L, 14L, 18L, 20L, 28L, 31L, 36L,
44L, 45L, 8L, 10L, 18L, 28L, 42L, 2L, 7L, 39L, 7L, 39L, 3L, 4L,
42L, 5L, 42L, 6L, 21L, 1L, 10L, 22L, 28L, 46L, 9L, 10L, 14L,
28L, 46L, 10L, 28L, 48L, 25L, 23L, 32L, 33L, 40L, 43L, 24L, 3L,
18L, 24L, 28L, 31L, 36L, 42L, 18L, 27L, 28L, 31L, 36L, 45L, 18L,
24L, 27L, 28L, 42L, 16L, 16L, 18L, 18L, 18L, 29L, 23L, 39L, 39L,
40L, 1L, 12L, 47L, 3L, 18L, 20L, 28L, 31L, 36L, 38L, 42L, 5L,
18L, 20L, 27L, 28L, 31L, 36L, 38L, 41L, 45L, 8L, 18L, 27L, 28L,
42L, 48L, 6L, 17L, 30L, 31L, 31L, 18L, 18L, 18L, 29L, 39L, 39L,
40L, 43L, 31L, 31L, 48L, 30L, 13L, 34L, 18L, 36L, 18L, 36L, 18L
), .Label = c("ER-11", "ER-19", "ER-21", "ER-22", "ER-29", "ER-30",
"ER-31", "ER-32", "ER-33", "ER-38", "ER-40", "ER-43", "ER-49",
"ER-8", "ER-AZ-04", "ER-AZ-05", "ER-AZ-06", "ER-AZ-07", "ER-AZ-08",
"ER-AZ-10", "ER-AZ-11", "ER-AZ-11=ER-47", "ER-AZ-13", "ER-AZ-14",
"ER-AZ-15", "ER-AZ-16", "ER-AZ-17", "ER-AZ-18", "ER-AZ-20", "ER-AZ-20=ER-27",
"ER-AZ-21", "ER-AZ-23", "ER-AZ-23=ER-52", "ER-AZ-24", "ER-AZ-29",
"ER-AZ-31", "ER-AZ-33", "ER-AZ-35", "ER-AZ-37", "ER-AZ-38", "ER-AZ-39",
"ER-AZ-40", "ER-AZ-43", "ER-AZ-44", "ER-AZ-45", "ER-AZ-49", "ER-AZ-51",
"ER-AZ-53"), class = "factor"), Freq = c(1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L)), class = c("grouped_df",
"tbl_df", "tbl", "data.frame"), row.names = c(NA, -161L), groups = structure(list(
Metastasis_Location = structure(c(1L, 1L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 4L, 4L, 4L, 4L, 4L, 4L, 5L, 5L, 5L, 5L, 5L, 5L,
5L, 5L, 6L, 6L, 6L, 6L, 6L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L,
7L, 7L, 8L, 8L, 9L, 9L, 9L, 10L, 10L, 10L, 10L, 10L, 10L,
10L, 10L, 10L, 11L, 11L, 11L, 11L, 11L), .Label = c("adrenal",
"bone", "breast", "liver", "lung", "muscle", "node", "pancreatic",
"peritoneum", "pleural", "skin"), class = "factor"), T0_T2_THERAPY_COD = structure(c(2L,
2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L,
2L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 1L, 1L, 2L, 2L, 2L, 2L, 1L,
1L, 1L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 2L, 2L, 2L, 1L, 1L, 1L,
1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L,
1L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L), .Label = c("A",
"F"), class = "factor"), T0_T2_PD_event = structure(c(2L,
2L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L,
2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L,
2L, 1L, 2L, 2L, 2L, 2L, 1L, 1L, 2L, 2L, 2L), .Label = c("No Progression",
"Progression"), class = "factor"), Gene_mut = structure(c(4L,
5L, 1L, 3L, 4L, 1L, 2L, 3L, 4L, 5L, 6L, 3L, 6L, 3L, 4L, 5L,
6L, 2L, 3L, 4L, 3L, 4L, 5L, 6L, 3L, 6L, 3L, 4L, 5L, 6L, 3L,
4L, 5L, 6L, 1L, 3L, 4L, 5L, 3L, 4L, 3L, 4L, 5L, 6L, 3L, 4L,
5L, 6L, 6L, 3L, 4L, 5L, 6L, 3L, 4L, 3L, 4L, 5L, 6L, 3L, 4L,
5L, 6L, 3L, 4L, 5L, 6L, 1L, 6L, 3L, 4L, 5L), .Label = c("AKT1",
"ERBB2", "ESR1", "PIK3CA", "TP53", "WT"), class = "factor"),
.rows = structure(list(1L, 2L, 3L, 4L, 5L, 6L, 7L, 8:12,
13:19, 20:22, 23L, 24L, 25:27, 28:35, 36:45, 46:50, 51L,
52L, 53L, 54:55, 56:58, 59L, 60L, 61L, 62L, 63L, 64:67,
68:72, 73:75, 76L, 77L, 78:79, 80L, 81L, 82L, 83:89,
90:95, 96:100, 101L, 102L, 103L, 104L, 105L, 106L, 107:108,
109L, 110L, 111:112, 113L, 114:121, 122:131, 132:137,
138:140, 141L, 142L, 143L, 144L, 145L, 146L, 147L, 148L,
149L, 150L, 151L, 152L, 153L, 154L, 155L, 156L, 157:158,
159:160, 161L), ptype = integer(0), class = c("vctrs_list_of",
"vctrs_vctr", "list"))), class = c("tbl_df", "tbl", "data.frame"
), row.names = c(NA, -72L), .drop = TRUE))
CodePudding user response:
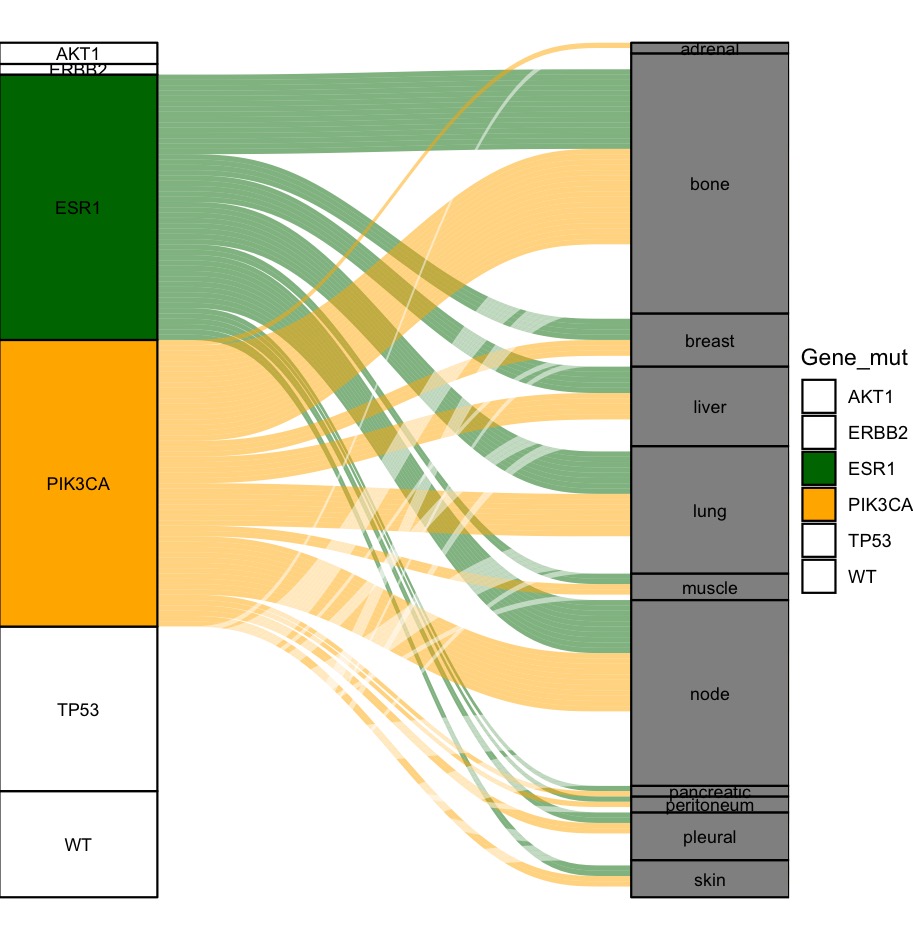
You're right to think of scale_fill_manual(). I think this is the more programmable alternative to passing a vector like colorfill to an aesthetic outside aes(). The following plot uses your data and color vector to control how the fill aesthetic is coded throughout the plot, and notice that fill is passed the same variable, Gene_mut, in both layers (alluvium and stratum):
ggplot(data = Allu,
aes(axis1 = Gene_mut, axis2 = Metastasis_Location, y = Freq))
geom_alluvium(aes(fill = Gene_mut),
curve_type = "quintic")
geom_stratum(aes(fill = Gene_mut), width = 1/4)
scale_fill_manual(values = colorfill)
geom_text(stat = "stratum", size = 3,
aes(label = after_stat(stratum)))
scale_x_discrete(limits = c("Metastasis_Location", "Gene_mut"),
expand = c(0.05, .05))
theme_void()
Since Metastasis_Location takes different values than Gene_mut, fill treats those strata as having missing values, which by default are colored grey. You can change that default by passing a color string to the na.value parameter of scale_fill_manual().