I'm trying to compare 2 columns of timestamps in a CSV file, and I only want to keep the rows where the date/time in column 1 is before the date/time in column 2. I'm not too sure where to start, since we're looking at comparing many different numbers (e.g. month, year, hour, minute, etc.) separately in relation with one another, including the AM/PM comparison.
Example: (date is [mm/dd/yyyy] format)
| 11/20/2018 3:00:13 PM | 11/23/2017 6:45:00 AM |
| 12/22/2019 4:00:12 PM | 1/10/2020 4:50:11 AM |
| 10/10/2018 2:02:19 PM | 10/07/2018 1:04:15 PM |
Here I would want to keep row 2 because the date in column 2 comes after the date in column 1, and I would not want to keep rows 1 & 3. Is there a neat way to do this in command line? (If not, any pointers to write a Python script would be very helpful)
Thanks in advance!
CodePudding user response:
In Python, you just need to convert each of the date values into datetime objects. They can then be easily compared with a simple < operator. For example:
from datetime import datetime
import csv
with open('input.csv') as f_input, open('output.csv', 'w', newline='') as f_output:
csv_input = csv.reader(f_input)
#header = next(csv_input)
csv_output = csv.writer(f_output)
#csv_output.writerow(header)
for row in csv_input:
date_col1 = datetime.strptime(row[0], '%m/%d/%Y %I:%M:%S %p')
date_col2 = datetime.strptime(row[1], '%m/%d/%Y %I:%M:%S %p')
if date_col1 < date_col2:
csv_output.writerow(row)
If your CSV file contains a header, uncomment the two lines. You can find more information on how the format string work with the 
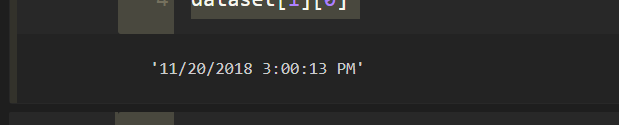
printing a single date from our dataset in order to check pattern :
Keep in mind that indexing in python starts with zero
dataset[1][0] # dataset[row][column]
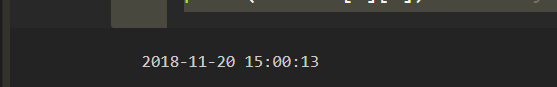
2.2) Pattern is month/day/year hour:min:sec AM/PM
pattern = "%m/%d/%Y %I:%M:%S %p"
successfully converted ^
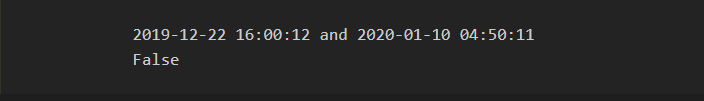
4) Now we will manually comparing dates in order to understand the concepts.
by simply using comparison operators we can compare the dates in python using datetime library
print(dataset[2][0] , "and" , dataset[2][1])
print(dataset[2][0] > dataset[2][1])
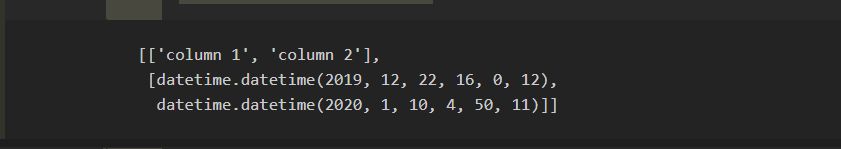
5) Now creating a separate list in which only those rows will be added where column 2's date is greater than column 1's date :
col2_greatorthan_col1 = []
adding heading in our new list :
col2_greatorthan_col1.append(["column 1" , "column 2"])
comparing each and every date and appending our desired row in our new list :
for i in dataset[1:]:
if i[1] > i[0]: # means if column 2's date is greater than column 1's date
col2_greatorthan_col1.append(i) # appending the filtered rows in our new list
col2_greatorthan_col1
6) Now simply creating a real world csv file which will have the same data as col2_greatorthan_col1
import csv
with open("new_dates.csv" , "w" , newline = "") as file :
writer = csv.writer(file)
writer.writerows(lst)
Result :
A new csv file by the name of new_dates.csv will be created in the same directory as your python code file. This file will only contain those rows where column 2's date is greater than column 1's date.
CodePudding user response:
I have saved the sample you provided in a tab separated file - with no headers. I have imported it as a DataFrame using (note that I specified your date format in date_parser):
import pandas as pd
import datetime as dt
df = pd.read_csv(PATH_TO_YOUR_FILE, sep="\t", names=["col1", "col2"], parse_dates=[0,1], date_parser=lambda x:dt.datetime.strptime(x, "%m/%d/%Y %I:%M:%S %p")
To select the rows you need:
df.loc[df.loc[:,"col2"]>df.loc[:,"col1"],:]
CodePudding user response:
With comman line tools you can use awk:
to convert 1st date to epoch format:
echo "11/20/2018 3:00:13 PM" |gawk -F'[/:]' '{print mktime($3" "$1" "$2" "$4" "$5" "$6" "$7)}'
same for the second field. And then subtract column 2 from column 1. If the result is positive this mean column 1 is after column 2
CodePudding user response:
You can use pd.to_datetime to parse the date-time strings and then use their comparison as a condition to filter the required rows.
Demo:
import pandas as pd
df = pd.DataFrame({
'start': ['11/20/2018 3:00:13 PM', '12/22/2019 4:00:12 PM', '10/10/2018 2:02:19 PM'],
'end': ['11/23/2017 6:45:00 AM', '1/10/2020 4:50:11 AM', '10/07/2018 1:04:15 PM']
})
result = pd.DataFrame(df[
pd.to_datetime(df['start'], format='%m/%d/%Y %I:%M:%S %p') <
pd.to_datetime(df['end'], format='%m/%d/%Y %I:%M:%S %p')
])
print(result)
Output:
start end
1 12/22/2019 4:00:12 PM 1/10/2020 4:50:11 AM
CodePudding user response:
Using Pandas
import pandas as pd
# Read tab delimited CSV file without header
# Names columns date1, date2
df = pd.read_csv("dates.csv",
header = None,
sep='\t',
parse_dates = [0, 1], # use default date parser i.e. parser.parser
names=["date1", "date2"])
# Filter (keep) row when date2 > date1
df = df[df.date2 > df.date1]
# Output to filtered CSV file using the original date format
df.to_csv('filtered_dates.csv', index = False, header = False, sep = '\t', date_format = "%Y/%m/%d %I:%M:%S %p")