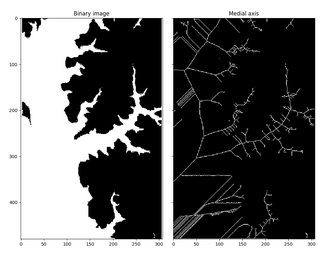
I am trying to make a skeleton of an image using medial_axis but for some reason, the result comes out really messy
does anyone have an idea how could I improve it?
I also tried using skeletonize and the skeleton itself seems better but it isn't quite what I'm looking for.
import matplotlib.pyplot as plt
from skimage.io import imread
from skimage.filters import threshold_otsu
from skimage.morphology import medial_axis
def Get_binary(image):
thresh = threshold_otsu(image)
binary = image > thresh
return binary
def Skeletonize_img():
im = Get_binary(imread('image_bin.png'))
im_ax = medial_axis(im)
fig, axes = plt.subplots(nrows=1, ncols=2, sharex=True, sharey=True,
figsize=(8,8))
ax = axes.ravel()
ax[0].imshow(im, cmap=plt.cm.gray)
ax[0].set_title('binary image')
ax[1].imshow(im_ax, cmap=plt.cm.gray)
ax[1].set_title('medial axis')
plt.tight_layout()
plt.show()
Skeletonize_img()
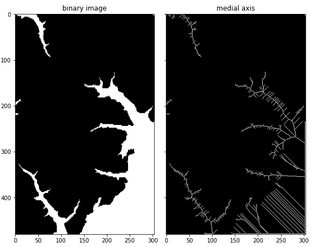
CodePudding user response:
from skimage import img_as_bool, io, color, morphology
import matplotlib.pyplot as plt
image = img_as_bool(color.rgb2gray(io.imread('image_bin.png')))
out = morphology.medial_axis(image)
f, (ax0, ax1) = plt.subplots(1, 2)
ax0.imshow(image, cmap='gray', interpolation='nearest')
ax1.imshow(out, cmap='gray', interpolation='nearest')
plt.show()
Also adding black border around the image will help