I am having an issue creating an example ODE given in Matlab to work with scipy's solve_ivp. In Matlab, the function is defined as
function fixed_point_linear_center()
clc; clf;
stepsize=.5;
xmin=-5;
xmax=5;
ymin=-5;
ymax=5;
[x,y] = meshgrid(xmin:stepsize:xmax,ymin:stepsize:ymax);
A = [0 1;-1 0];
dx = A(1,1)*x A(1,2)*y;
dy = A(2,1)*x A(2,2)*y;
% Strange scaling for nicer output, only "cosmetics"
eunorm = ( dx.^2 dy.^2 ).^(0.35);
dx = dx./eunorm;
dy = dy./eunorm;
quiver(x,y,dx,dy);
axis([xmin xmax ymin ymax]);
grid on; xlabel('x'); ylabel('y');
tspan=[0 100];
x0stepsize=0.25;
for x0=xmin:x0stepsize:xmax
hold on
ic = [x0 0];
[~,x] = ode45(@(t,x) f(t,x,A),tspan,ic);
plot(x(:,1),x(:,2),'r');
hold on
ic = [0 x0];
[~,x] = ode45(@(t,x) f(t,x,A),tspan,ic);
plot(x(:,1),x(:,2),'r');
end
hold off
end
function dx = f(~,x,A)
dx = A*[x(1); x(2)];
end
to calculate the solution which looks like this
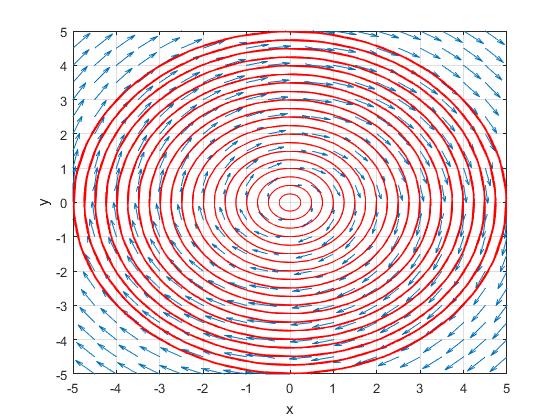
, however if I recreate the functions in python like this
def fixed_point_linear_center():
stepsize = 0.5
x0stepsize = 0.25
xmin = -5
xmax = 5
ymin = -5
ymax = 5
x = np.arange(xmin, xmax stepsize, stepsize)
xval = np.arange(xmin, xmax x0stepsize, x0stepsize)
y = np.arange(ymin, ymax stepsize, stepsize)
yval = np.arange(ymin, ymax stepsize*0.25, stepsize*0.25) # evaluate 4 times for smoothness
[X, Y] = np.meshgrid(x, y)
A = np.array([[0,1],[-1,0]])
dx = A[0,0]*X A[0,1]*Y # 21x21
dy = A[1,0]*X A[1,1]*Y # 21x21
f = lambda t,x,A : np.dot(A,[[x[0]],[x[1]]])
# Strange scaling for nicer output, but only "cosmetics"
eunorm = np.float_power(( dx**2 dy**2 ), 0.35) #( dx**2 dy**2 )**0.35
eunorm[10,10] = 0.001 # center is 0 which violates division
dx = dx/eunorm
dy = dy/eunorm
plt.figure(figsize = (15,12))
plt.quiver(X, Y, dx, dy, angles = 'xy', color='#0086b3', width=0.0015)
plt.grid()
plt.xlabel('x')
plt.ylabel('y')
plt.axis([xmin,xmax,ymin,ymax])
tspan=[0,100]
for x0 in xval:
ic = [x0,0]
#[~,x] = ode45(@(t,x) f(t,x,A),tspan,ic);
solution = solve_ivp(f, [xmin, xmax], ic, method='RK45', t_eval=yval, dense_output=True, args=(A,))
#solution = solve_ivp(f, [xmin, xmax], [x0], method='RK45', t_eval=yval, dense_output=False, args=(0,A))
#solution = solve_ivp(f, [tmin, tmax], [ic], method='RK45', t_eval=tval, args=(A), dense_output=False)
plt.plot(solution.y[1], solution.y[0],'r')
fixed_point_linear_center()
I get errors like
ValueError: shapes (2,2) and (2,1,2) not aligned: 2 (dim 1) != 1 (dim 1)
or similar, depending on what I already tried to rewrite f to. As I understand, solve_ivp expects a single value in the x0 array, while I return a 2x1 vector. It also doesn't accept a vector as value in it's x0 array like [[x0,0]]
Now I wonder if scipy.solve_ivp able to do the calculation like ode45 for parameter space (and how do I do it) or do I have to do the calculation otherwise?
(I checked already, that all other matrices and return values are identical with the matlab calculation.)
[EDIT 2]
okay, it works now. The plot parameter for x had to be solution.y[1] of course!
CodePudding user response:
Just like the Matlab solver, solve_ivp expects the state to be a single vector. Change
f = lambda t,x1,x2, A : np.dot(A,[[x1],[x2]])
to
f = lambda t, x, A : np.dot(A, x)
Also, to ensure ensure that solve_ivp interprets the shape of the argument A correctly, pass it to solve_ivp with args=(A,) (note the use of the comma).
