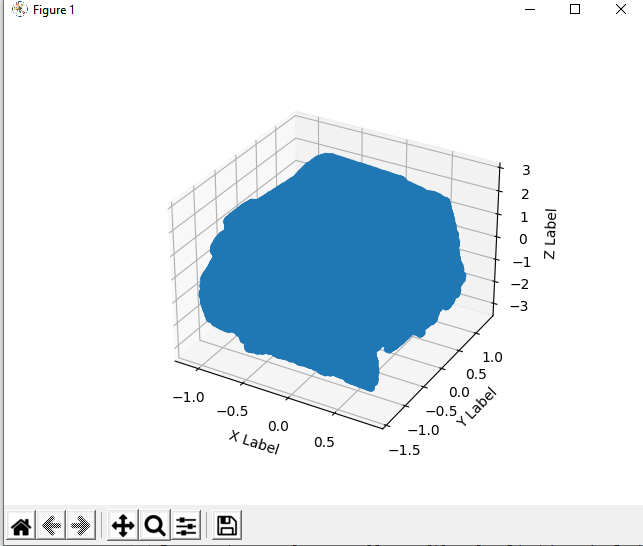
I am trying to plot a small xyz file of a 3d object in Python. Unfortunately it's a little too thick, see below:
Code below is what generated it:
input_file = sys.argv[1];
print("input_file :: " str(input_file))
data = pd.read_csv(input_file, header=None)
data = data.to_numpy();
print("----- Data read successfully. Shape and random row: ------")
print(data.shape)
print(data[123,:])
fig = plt.figure()
ax = fig.add_subplot(projection='3d')
ax.scatter(data[:,0], data[:,1], data[:,2], linewidth=1)
ax.set_xlabel('X Label')
ax.set_ylabel('Y Label')
ax.set_zlabel('Z Label')
plt.show()
Data is of shape (316369, 3) and each row's example data looks something like: [-0.97320002 0.44980001 0.79809999]
Here's how I'd like it look, referencing one of my 3d programs:
Are there Matplotlib settings which would let me approach this?
CodePudding user response:
Try to change the marker and s (size) parameters of scatter:
ax.scatter(data[:,0], data[:,1], data[:,2], marker='.', s=1)
You likely have to go through trial and error to find a nice value for s (you can use decimal values). Know that it is not linear but a squared value.
CodePudding user response:
To add to @mozway's answer, setting the transparency of your markers can also help. This can be accomplished with the alpha parameter, or by specifying an RGBA color value explicitly with a <1 alpha (preferably with the color keyword argument, though c works as well).