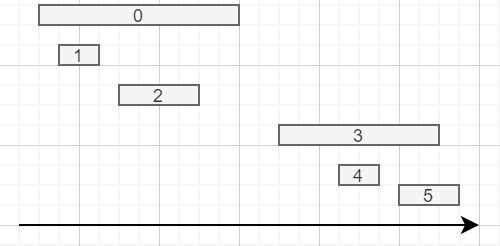
I have a DataFrame like this (but much larger):
id start end
0 10 20
1 11 13
2 14 18
3 22 30
4 25 27
5 28 31
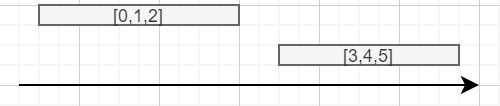
I am trying to efficiently merge overlapping intervals in PySpark, while saving in a new column 'ids', which intervals were merged, so that it looks like this:
start end ids
10 20 [0,1,2]
22 31 [3,4,5]
Visualisation:
from:
Can I do this without using an udf?
edit: the order of id and start are not necessarily the same.
CodePudding user response:
You can use window function to compare previous rows with current row, to build a column that determine if current row is the start of a new interval, then sum over this column to build a interval id. Then you group by this interval id to get your final dataframe.
If you call input_df your input dataframe, the code will be as follows:
from pyspark.sql import Window
from pyspark.sql import functions as F
all_previous_rows_window = Window \
.orderBy('start') \
.rowsBetween(Window.unboundedPreceding, Window.currentRow)
result = input_df \
.withColumn('max_previous_end', F.max('end').over(all_previous_rows_window)) \
.withColumn('interval_change', F.when(
F.col('start') > F.lag('max_previous_end').over(Window.orderBy('start')),
F.lit(1)
).otherwise(F.lit(0))) \
.withColumn('interval_id', F.sum('interval_change').over(all_previous_rows_window)) \
.drop('interval_change', 'max_previous_end') \
.groupBy('interval_id') \
.agg(
F.collect_list('id').alias('ids'),
F.min('start').alias('start'),
F.max('end').alias('end')
).drop('interval_id')
So you can merge your intervals without any user-defined function. However, every time we use a window, code is executed on only on one executor, as our windows don't have partitions.