Dataframe was extracted to a temp table to plot the data density per time unit (1 day):
val dailySummariesDf =
getDFFromJdbcSource(SparkSession.builder().appName("test").master("local").getOrCreate(), s"SELECT * FROM values WHERE time > '2020-06-06' and devicename='Voltage' limit 100000000")
.persist(StorageLevel.MEMORY_ONLY_SER)
.groupBy($"digital_twin_id", window($"time", "1 day")).count().as("count")
.withColumn("windowstart", col("window.start"))
.withColumn("windowstartlong", unix_timestamp(col("window.start")))
.orderBy("windowstart")
dailySummariesDf.
registerTempTable("bank")
Then I plot it with %sql processor
%sql
select windowstart, count
from bank
and
%sql
select windowstartlong, count
from bank
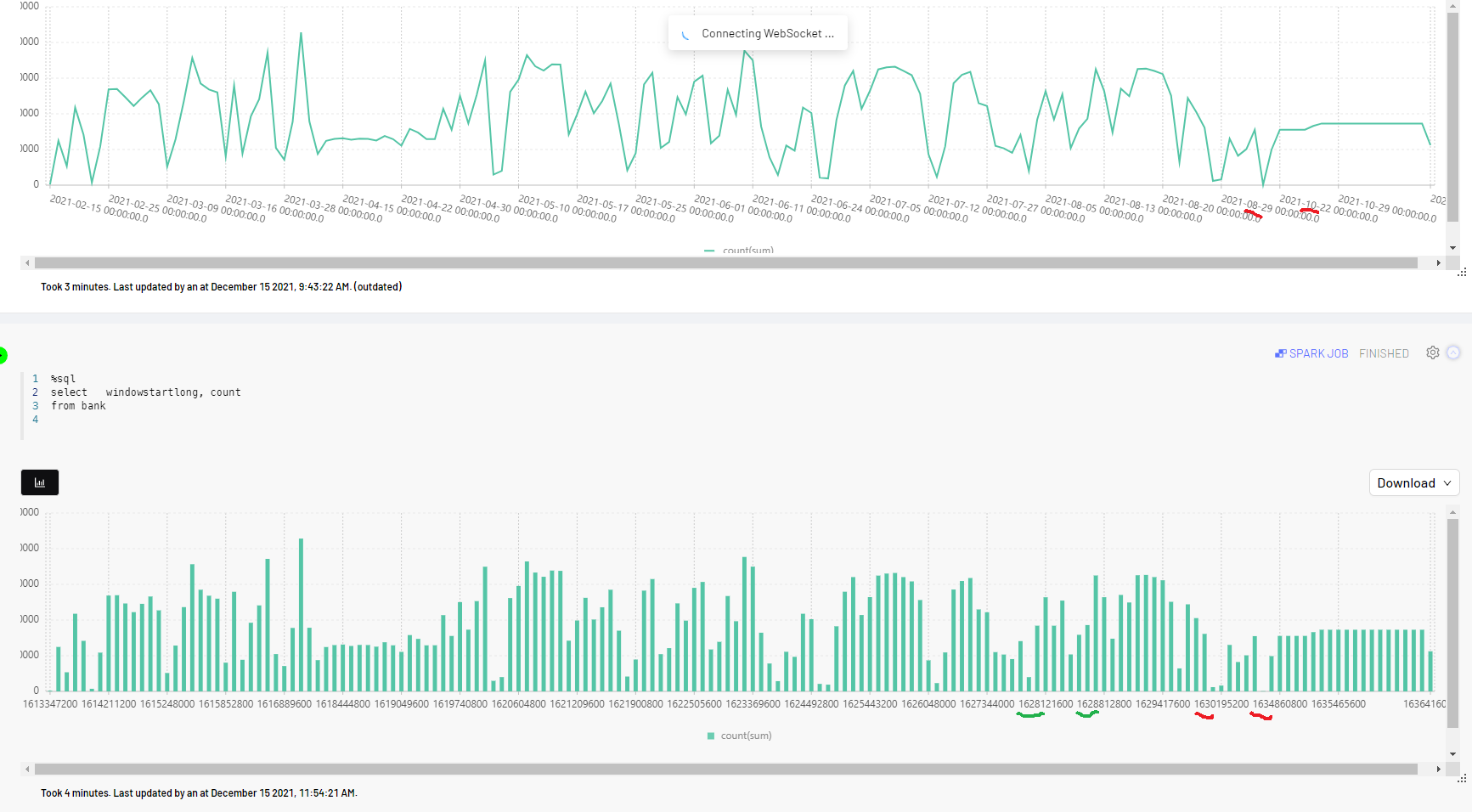
What I get is shown below:
So, my expectation is to have gaps in this graph, as there were days with no data at all. But instead I see it being plotted densely, with October days plotted right after August, not showing a gap for September.
How can I force those graphs to display gaps and regard the real X axis values?
CodePudding user response:
Indeed, grouping a dataset by window column won't produce any rows for the intervals that did not contain any original rows within those intervals.
One way to deal with that I can think of, is to add a bunch of fake rows ("manually fill in the gaps" in raw dataset), and only then apply a groupBy/window. For your case, that can be done by creating a trivial one-column dataset containing all the dates within a range you're interested in, and then joining it to your original dataset.
Here is my quick attempt:
import spark.implicits._
import org.apache.spark.sql.types._
// Define sample data
val df = Seq(("a","2021-12-01"),
("b","2021-12-01"),
("c","2021-12-01"),
("a","2021-12-02"),
("b","2021-12-17")
).toDF("c","d").withColumn("d",to_timestamp($"d"))
// Define a dummy dataframe for the range 12/01/2021 - 12/30/2021
import org.joda.time.DateTime
import org.joda.time.format.DateTimeFormat
val start = DateTime.parse("2021-12-01",DateTimeFormat.forPattern("yyyy-MM-dd")).getMillis/1000
val end = start 30*24*60*60
val temp = spark.range(start,end,24*60*60).toDF().withColumn("tc",to_timestamp($"id".cast(TimestampType))).drop($"id")
// Fill the gaps in original dataframe
val nogaps = temp.join(df, temp.col("tc") === df.col("d"), "left")
// Aggregate counts by a tumbling 1-day window
val result = nogaps.groupBy(window($"tc","1 day","1 day","5 hours")).agg(sum(when($"c".isNotNull,1).otherwise(0)).as("count"))
result.withColumn("windowstart",to_date(col("window.start"))).select("windowstart","count").orderBy("windowstart").show(false)
----------- -----
|windowstart|count|
----------- -----
|2021-12-01 |3 |
|2021-12-02 |1 |
|2021-12-03 |0 |
|2021-12-04 |0 |
|2021-12-05 |0 |
|2021-12-06 |0 |
|2021-12-07 |0 |
|2021-12-08 |0 |
|2021-12-09 |0 |
|2021-12-10 |0 |
|2021-12-11 |0 |
|2021-12-12 |0 |
|2021-12-13 |0 |
|2021-12-14 |0 |
|2021-12-15 |0 |
|2021-12-16 |0 |
|2021-12-17 |1 |
|2021-12-18 |0 |
|2021-12-19 |0 |
|2021-12-20 |0 |
----------- -----
For illustration purposes only :)