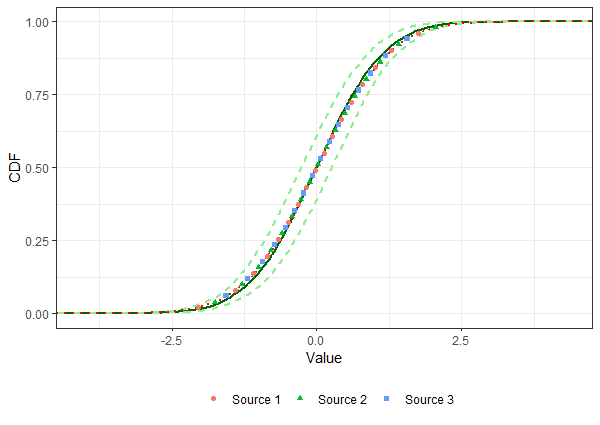
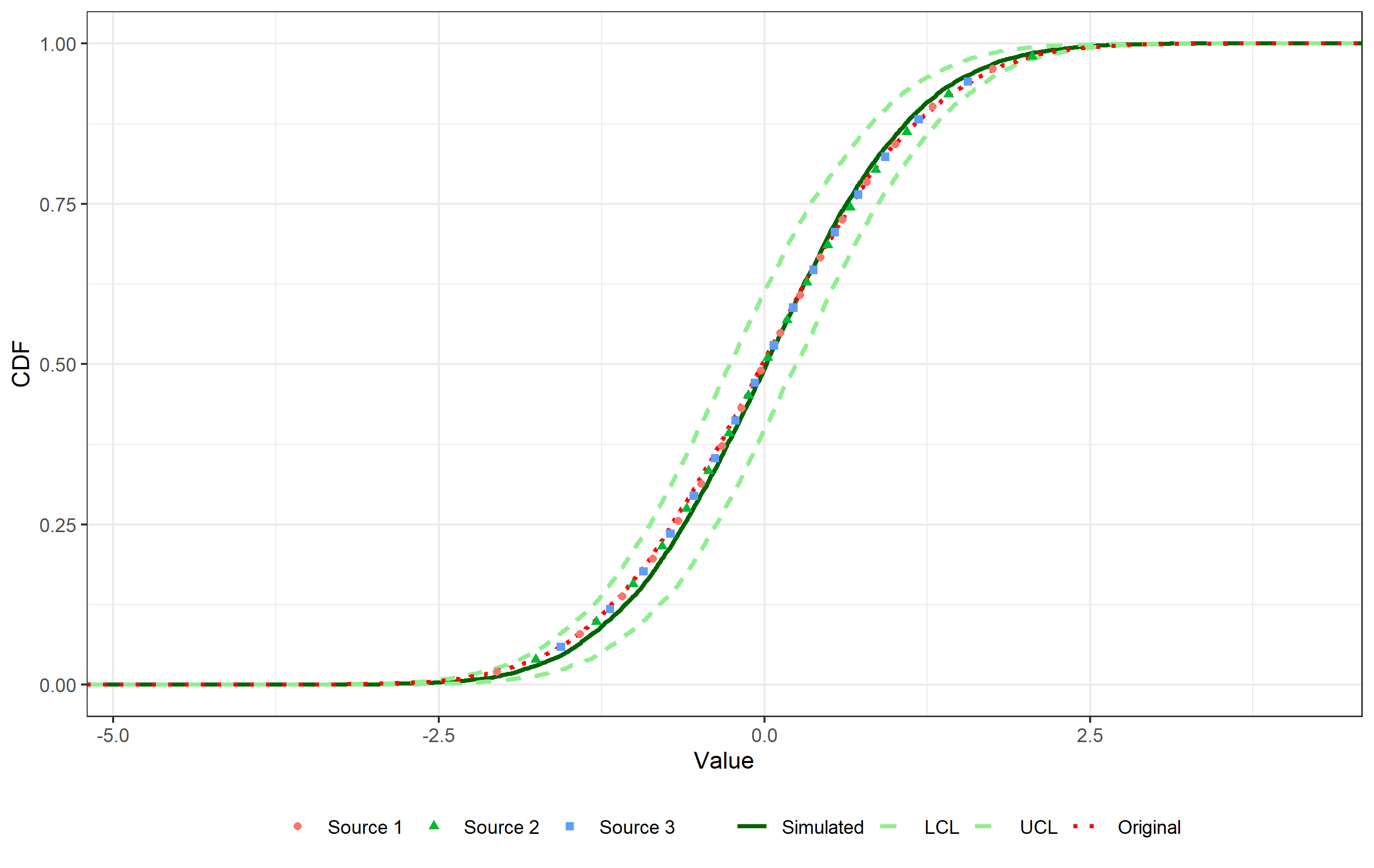
After estimating the parameters of a distribution through bootstrapping, starting from "known" data (i.e., coming from the literature, experiments, and so on) for wihch the teoretical distribution is known, I want to display on the same plot the original data (with associated cumulative probability), and the CDFs of both the "true" distribution and the estimated one.
However, I am failing to display data and curves in the same legend.
Minimal reproducible example:
libary(ggplot2)
n.studies <- 50
cumulative.prob <- seq(1 / n.studies, 1 - 1 / n.studies, length.out = n.studies)
known.values <- qnorm(cumulative.prob)
group <- rep(c("Source 1", "Source 2", "Source 3"), length.out = n.studies)
data <- data.frame(x = known.values, y = cumulative.prob, group = group)
# Simulate the bootstrap procedure
sampler <- function(x) {
bootstrap <- sample(x = known.values, size = length(known.values), replace = T)
return(c(mean = mean(bootstrap), sd = sd(bootstrap)))
}
simulated.statistics <- as.data.frame(do.call(rbind, lapply(rep(1, 10000), sampler)))
simulated.dist <- data.frame("Simulated" = rnorm(10000, mean = mean(simulated.statistics$mean), sd = mean(simulated.statistics$sd)),
"LCL" = rnorm(10000, mean = quantile(simulated.statistics$mean, .025, names = F), sd = mean(simulated.statistics$sd)),
"UCL" = rnorm(10000, mean = quantile(simulated.statistics$mean, .975, names = F), sd = mean(simulated.statistics$sd)),
"Original" = rnorm(10000))
# Plot data and CDFs
g <- ggplot()
stat_ecdf(data = simulated.dist, aes(x = Simulated), colour = "darkgreen", linetype = "solid", geom = "step", lwd = 1)
stat_ecdf(data = simulated.dist, aes(x = LCL), colour = "lightgreen", linetype = "dashed", geom = "step", lwd = 1)
stat_ecdf(data = simulated.dist, aes(x = UCL), colour = "lightgreen", linetype = "dashed", geom = "step", lwd = 1)
stat_ecdf(data = simulated.dist, aes(x = Original), colour = "red", linetype = "dotted", geom = "step", lwd = 1)
geom_point(data = data, aes(x = x, y = y, colour = group, shape = group))
xlab("Value")
ylab("CDF")
theme_bw()
theme(legend.position = "bottom",
legend.title = element_blank())
print(g)
What I want to obtain is exactly this graph, with a legend displaying in addition to the data points (exactly as they are) the four curves (i.e. two "dashed lightgreen" curves labeled as "LCL" and "UCL", a "solid darkgreen" curve labeled "Simulated", and a "red dotted" curve labeled "Original"). So far I tried to manually set legends for colors, shapes and linetypes, but I am either obtaining "more than one legend" or an incorrect graph (e.g., points or curves are disappearing), I suppose this is due to the fact that stat_ecdf and geom_point do not share the same aesthetics.
Thank you in advance for your precious help
CodePudding user response: