In my example I have a 3D point cloud and want to find the outline for each z-layer. My current approach is the following:
library(rgl) #just for 3D visualisation purposes of the cube
cube = data.frame(x = rep(1:10,1000),
y = rep(1:10, 100, each = 10),
z = rep(1:10,100,each = 100)) #3D point cloud
xyz_list = split(cube, cube[,3]) #split into layers by unique z-values
t0 = Sys.time()
outline = lapply(xyz_list, function(k){
xmax = merge(aggregate(y ~ x, FUN = max, data = k), k) #maximum y-value for each unique x-value
xmin = merge(aggregate(y ~ x, FUN = min, data = k), k) #minimum y-value for each unique x-value
ymax = merge(aggregate(x ~ y, FUN = max, data = k), k) #maximum x-value for each unique y-value
ymin = merge(aggregate(x ~ y, FUN = min, data = k), k) #minimum x-value for each unique y-value
mm = rbind(xmax,xmin,ymax,ymin) #collect all minimum and maximum values
mm = mm[!duplicated(mm),] #remove duplicate rows
})
t1 = Sys.time()
print(t1 - t0)
outline = do.call(rbind,outline)#merge lists
plot3d(cube)
plot3d(outline, col = "red", add = TRUE, size = 5)
Which takes approx. 0.33 secs
Now I thought about passing the dataframe (xyz_list) to a function named of outside of lapply and move all the code from inside lapply to that function as I need to repeat the operations several times later on:
of = function(df, dim1, dim2){
xmax = merge(aggregate(df[,dim1] ~ df[,dim2], FUN = max, data = df), df)
xmin = merge(aggregate(df[,dim1] ~ df[,dim2], FUN = min, data = df), df)
ymax = merge(aggregate(df[,dim2] ~ df[,dim1], FUN = max, data = df), df)
ymin = merge(aggregate(df[,dim2] ~ df[,dim1], FUN = min, data = df), df)
mm = rbind(xmax,xmin,ymax,ymin) #collect all minimum and maximum values
mm = mm[!duplicated(mm),] #remove duplicate rows
return(mm)
}
t0 = Sys.time()
outline = lapply(xyz_list, function(k){
mm = of(k, 2, 1)
})
t1 = Sys.time()
print(t1 - t0)
Which takes about 13 secs.
I don't understand why my code has become so much slower in the second example. Is there some way to make the function of more efficient?
CodePudding user response:
# dummy data
cube <- data.frame(x = rep(1:10,1000),
y = rep(1:10, 100, each = 10),
z = rep(1:10,100,each = 100)
)
# split into list
xyz_list <- split(cube, cube[,3])
op's original method (lapply only)
outline <- lapply(xyz_list, function(k)
{
xmax = merge(aggregate(y ~ x, FUN = max, data = k), k) #maximum y-value for each unique x-value
xmin = merge(aggregate(y ~ x, FUN = min, data = k), k) #minimum y-value for each unique x-value
ymax = merge(aggregate(x ~ y, FUN = max, data = k), k) #maximum x-value for each unique y-value
ymin = merge(aggregate(x ~ y, FUN = min, data = k), k) #minimum x-value for each unique y-value
mm = rbind(xmax,xmin,ymax,ymin) #collect all minimum and maximum values
mm = mm[!duplicated(mm),] #remove duplicate rows
})
op's attempt to create function (then lapply)
of <- function(df, dim1, dim2)
{
xmax = merge(aggregate(df[,dim1] ~ df[,dim2], FUN = max, data = df), df)
xmin = merge(aggregate(df[,dim1] ~ df[,dim2], FUN = min, data = df), df)
ymax = merge(aggregate(df[,dim2] ~ df[,dim1], FUN = max, data = df), df)
ymin = merge(aggregate(df[,dim2] ~ df[,dim1], FUN = min, data = df), df)
mm = rbind(xmax,xmin,ymax,ymin) #collect all minimum and maximum values
mm = mm[!duplicated(mm),] #remove duplicate rows
return(mm)
}
new, improved function
of1 <- function(df, y, x)
{
y_x <- as.formula(paste(y, '~', x))
x_y <- as.formula(paste(x, '~', y))
xmax = merge(aggregate(y_x, FUN = max, data = df), df)
xmin = merge(aggregate(y_x, FUN = min, data = df), df)
ymax = merge(aggregate(x_y, FUN = max, data = df), df)
ymin = merge(aggregate(x_y, FUN = min, data = df), df)
mm = rbind(xmax,xmin,ymax,ymin) #collect all minimum and maximum values
mm = mm[!duplicated(mm),] #remove duplicate rows
return(mm)
}
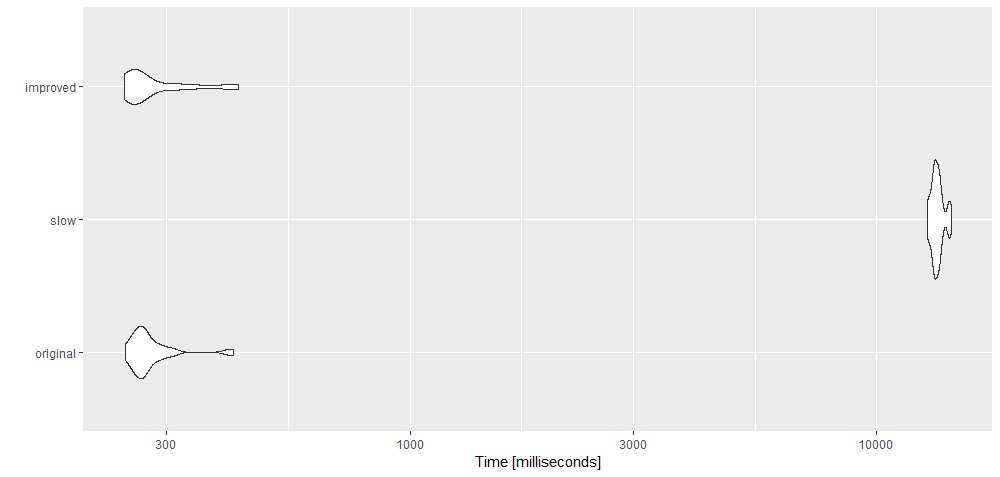
microbenchmark
library(microbenchmark)
library(ggplot2)
a <-
microbenchmark(original = {outline = lapply(xyz_list, function(k)
{
xmax = merge(aggregate(y ~ x, FUN = max, data = k), k) #maximum y-value for each unique x-value
xmin = merge(aggregate(y ~ x, FUN = min, data = k), k) #minimum y-value for each unique x-value
ymax = merge(aggregate(x ~ y, FUN = max, data = k), k) #maximum x-value for each unique y-value
ymin = merge(aggregate(x ~ y, FUN = min, data = k), k) #minimum x-value for each unique y-value
mm = rbind(xmax,xmin,ymax,ymin) #collect all minimum and maximum values
mm = mm[!duplicated(mm),] #remove duplicate rows
})}
, slow = {outline1 = lapply( xyz_list, function(k) { of(k, 2, 1) } )}
, improved = {outline2 = lapply( xyz_list, function(k) of1(k, 'y', 'x') )}
, times = 30
)
autoplot(a)
identity check
identical(outline, outline2)
[1] TRUE
CodePudding user response:
data.table solution:
I would recommend just subsetting on the larger data.table as needed instead of splitting it into separate data.tables by z layer. But if outline is really needed as a list of data.tables, we can split the z layers out after doing the summarizing:
library(data.table)
cube <- data.table(x = rep(1:10, 1000),
y = rep(1:10, 100, each = 10),
z = rep(1:10, 100, each = 100)) #3D point cloud
system.time({
nms <- c("x", "y", "z")
outline2 <- unique(rbindlist(lapply(1:2, function(i) setnames(cube[, .(range(.SD)), by = c(nms[-i]), .SDcols = nms[i]], "V1", nms[i])), use.names = TRUE))
setcolorder(outline2, nms)
outline2 <- split(outline2, outline2[[3]])
})
#> user system elapsed
#> 0.03 0.00 0.05
Compare to the original non-function solution:
system.time({
xyz_list <- split(cube, cube[,3]) #split into layers by unique z-values
outline1 <- lapply(xyz_list, function(k){
xmax <- merge(aggregate(y ~ x, FUN = max, data = k), k) #maximum y-value for each unique x-value
xmin <- merge(aggregate(y ~ x, FUN = min, data = k), k) #minimum y-value for each unique x-value
ymax <- merge(aggregate(x ~ y, FUN = max, data = k), k) #maximum x-value for each unique y-value
ymin <- merge(aggregate(x ~ y, FUN = min, data = k), k) #minimum x-value for each unique y-value
mm <- rbind(xmax,xmin,ymax,ymin) #collect all minimum and maximum values
mm <- mm[!duplicated(mm),] #remove duplicate rows
})
})
#> user system elapsed
#> 0.64 0.01 0.66
If a function that operates on a list of pre-split layers is really needed:
of <- function(dt, dim1, dim2) {
setcolorder(unique(rbindlist(lapply(c(dim1, dim2), function(i) setnames(dt[, .(range(.SD)), by = c(nms[-i]), .SDcols = nms[i]], "V1", nms[i])), use.names = TRUE)), nms)
}
system.time({
outline3 <- lapply(xyz_list, function(k) of(k, 1, 2))
})
#> user system elapsed
#> 0.06 0.00 0.06
We'll verify that the solutions all return the same set of values. In order to compare, we need to convert the outline1 data.frames to data.tables and reset their rownames. We also sort all the data.tables.
for (i in 1:length(outline1)) {
setorder(setDT(outline1[[i]]))
setorder(outline2[[i]])
setorder(outline3[[i]])
rownames(outline1[[i]]) <- NULL
}
identical(outline1, outline2)
#> [1] TRUE
identical(outline1, outline3)
#> [1] TRUE
Created on 2022-01-31 by the reprex package (v2.0.1)