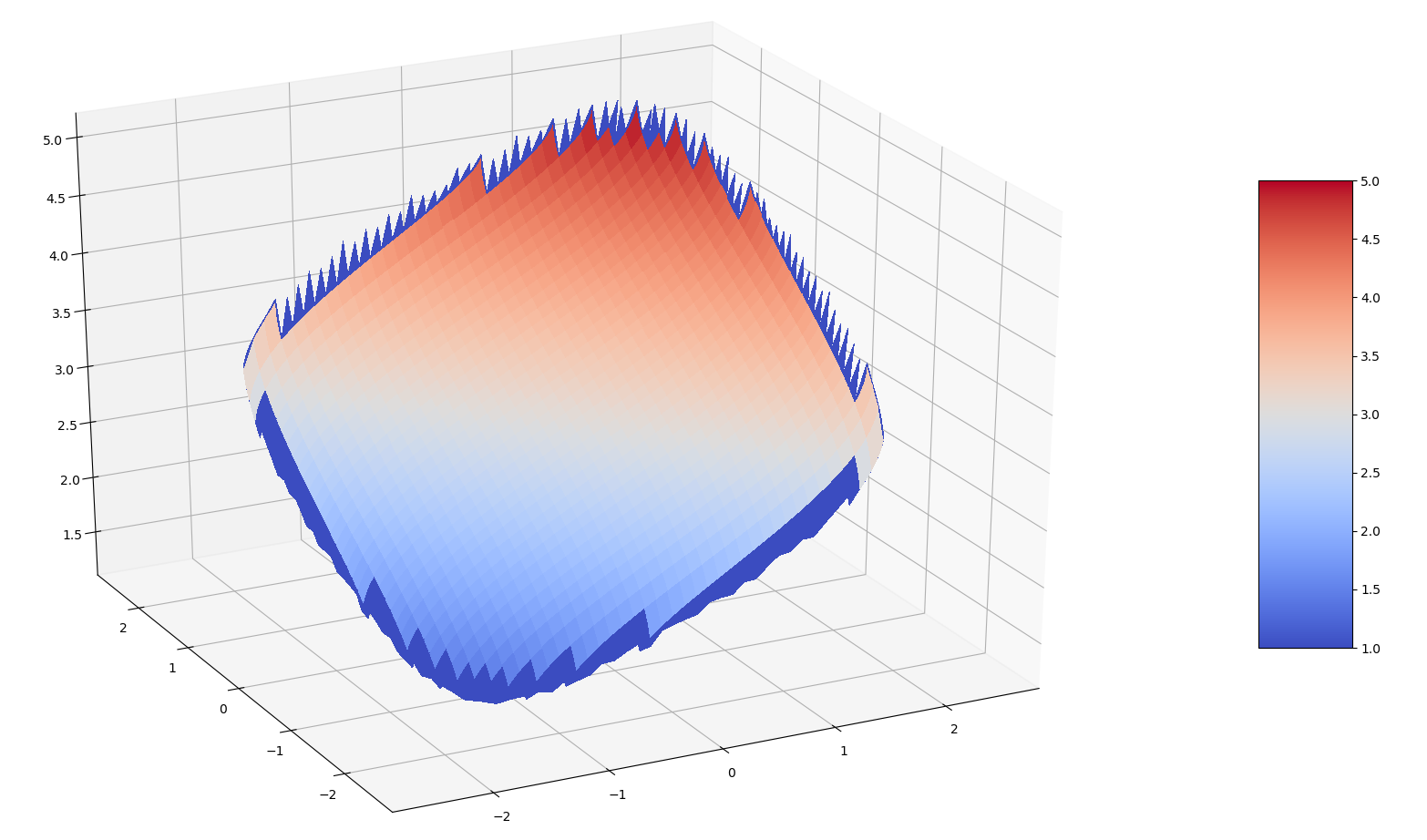
I would like to make surface plot of a function which is discontinuous at certain values in parameter space. It is near these discontinuities that the plot's coloring becomes incorrect, as shown in the picture below. How can I fix this?
My code is given below:
from mpl_toolkits.mplot3d import Axes3D
import matplotlib.pyplot as plt
from matplotlib import cm
import numpy as np
def phase(mu_a, mu_b, t, gamma):
theta = 0.5*np.arctan2(2*gamma, mu_b-mu_a)
epsilon = 2*gamma**2/np.sqrt((mu_a-mu_b)**2 4*gamma**2)
y1 = np.arccos(0.5/t*(-mu_a*np.sin(theta)**2 -mu_b*np.cos(theta)**2 - epsilon))
y2 = np.arccos(0.5/t*(-mu_a*np.cos(theta)**2 -mu_b*np.sin(theta)**2 epsilon))
return y1 y2
fig = plt.figure()
ax = fig.gca(projection='3d')
# Make data.
X = np.arange(-2.5, 2.5, 0.01)
Y = np.arange(-2.5, 2.5, 0.01)
X, Y = np.meshgrid(X, Y)
Z = phase(X, Y, 1, 0.6)
# Plot the surface.
surf = ax.plot_surface(X, Y, Z, cmap=cm.coolwarm, linewidth=0, antialiased=False)
surf.set_clim(1, 5)
fig.colorbar(surf, shrink=0.5, aspect=5)
plt.show()
CodePudding user response:
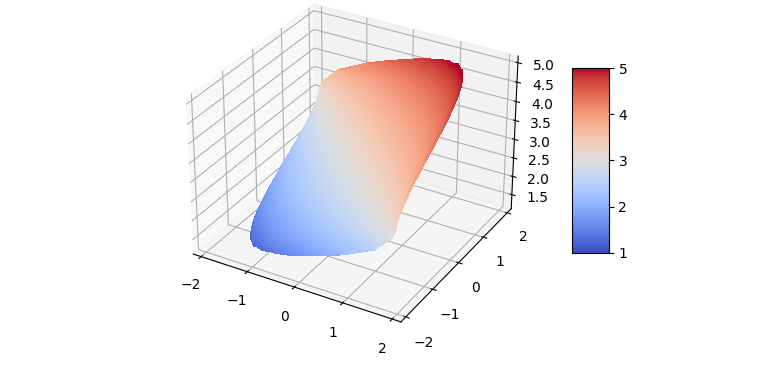
An idea is to make all the arrays 1D, filter out the NaN values and then call ax.plot_trisurf:
from mpl_toolkits.mplot3d import Axes3D
import matplotlib.pyplot as plt
from matplotlib import cm
import numpy as np
def phase(mu_a, mu_b, t, gamma):
theta = 0.5 * np.arctan2(2 * gamma, mu_b - mu_a)
epsilon = 2 * gamma ** 2 / np.sqrt((mu_a - mu_b) ** 2 4 * gamma ** 2)
with np.errstate(divide='ignore', invalid='ignore'):
y1 = np.arccos(0.5 / t * (-mu_a * np.sin(theta) ** 2 - mu_b * np.cos(theta) ** 2 - epsilon))
y2 = np.arccos(0.5 / t * (-mu_a * np.cos(theta) ** 2 - mu_b * np.sin(theta) ** 2 epsilon))
return y1 y2
fig = plt.figure()
ax = fig.add_subplot(projection='3d')
# Make data.
X = np.linspace(-2.5, 2.5, 200)
Y = np.linspace(-2.5, 2.5, 200)
X, Y = np.meshgrid(X, Y)
X = X.ravel() # make the array 1D
Y = Y.ravel()
Z = phase(X, Y, 1, 0.6)
mask = ~np.isnan(Z) # select the indices of the valid values
# Plot the surface.
surf = ax.plot_trisurf(X[mask], Y[mask], Z[mask], cmap=cm.coolwarm, linewidth=0, antialiased=False)
surf.set_clim(1, 5)
fig.colorbar(surf, shrink=0.5, aspect=5)
plt.show()
Some remarks:
plot_trisurfwill join the XY-values via triangles; this only works well if the domain is convex- to make things draw quicker, less points could be used (the original used 500x500 points, the code here reduces that to 200x200
- calling
fig.gca(projection='3d')has been deprecated; instead, you could callfig.add_subplot(projection='3d') - the warnings for dividing by zero or using arccos out of range can be temporarily suppressed; that way the warning will still be visible for situations when such isn't expected behavior