I am running the same piece of code, with the same seed, same package versions, same R version, on 3 different systems: 1) my computer 2) a linux cluster and 3) 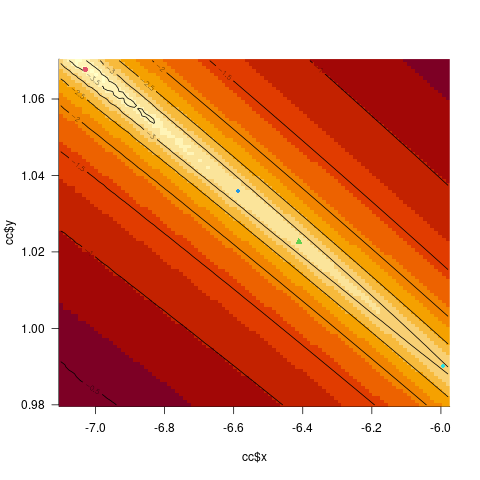
[red: Linux, green: Windows/convergence failure, blue: Windows/perturbed start, cyan: Nelder-Mead]
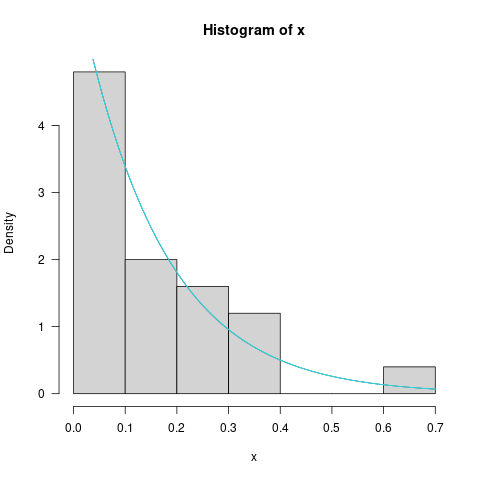
These are the corresponding fits to the distribution:
As you can see (or probably can't!), all the fits are basically identical. If you calculate the negative log-likelihood, you'll see that they differ by less than 0.001 units [i.e., negligibly]. (You can also tell this because all the points in the first image lie within the log10(difference) = -3 contour.)
So the differences between answers don't really matter, just the annoyance of getting an error. You could (1) use a while loop try() to perturb the starting value a little bit until you get an answer; (2) drop the bounds to allow Nelder-Mead to work: (3) use bbmle or some other tool that lets you be a little bit more robust/defensive about the optimization procedure ...
nllfun <- function(mu, sigma) {
-sum(log(dtruncnorm(x, a = 0, b = 1, mean = mu, sd = sigma)))
}
library(emdbook)
library(truncnorm)
p1 <- c(-7.02938981, 1.06779942) ## Linux
p2 <- c(-6.411, 1.022651) ## Windows (convergence error)
p3 <- c(-6.587645, 1.0359466) ## Windows (perturbed start)
p4 <- c(-5.9937989, 0.9901366) ## Windows (Nelder-Mead/no bounds)
cc <- curve3d(nllfun(x,y), xlim = c(-7.1, -5.98), ylim = c(0.98, 1.07),
n = c(101, 101), sys3d = "none")
image(cc$x, cc$y, log10(cc$z-min(cc$z) 1e-4))
contour(cc$x, cc$y, log10(cc$z-min(cc$z) 1e-4), add = TRUE)
points(p1[1], p1[2], pch = 16, col = 2)
points(p2[1], p2[2], pch = 17, col = 3)
points(p3[1], p3[2], pch = 18, col = 4)
points(p4[1], p4[2], pch = 18, col = 5)
hist(x, freq=FALSE)
curve(dtruncnorm(x, a=0, b=1, mean=p1[1], sd = p1[2]), col = 2, add=TRUE)
curve(dtruncnorm(x, a=0, b=1, mean=p2[1], sd = p2[2]), col = 3, add=TRUE)
curve(dtruncnorm(x, a=0, b=1, mean=p3[1], sd = p3[2]), col = 4, add=TRUE)
curve(dtruncnorm(x, a=0, b=1, mean=p4[1], sd = p4[2]), col = 5, add=TRUE)