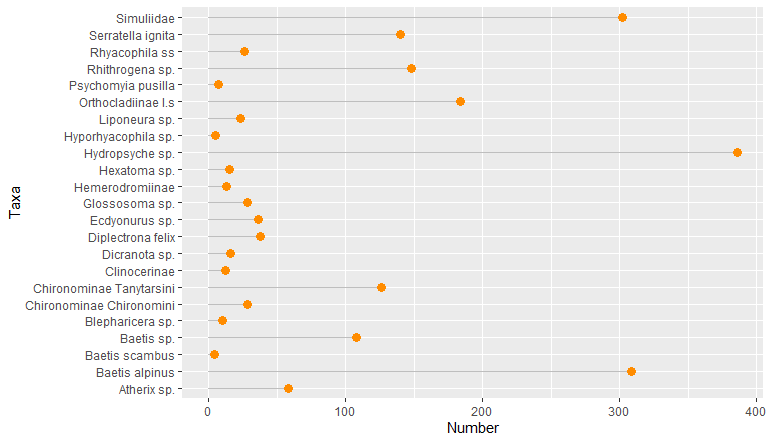
I am trying to plot data in ggplot and I would like them to appear in a certain order based on another column. I tried this on several types of chart (geom_bar, lollipop graph or pie chart) but the same problem keeps occuring so I guess I don't understand something.
I would like to plot the column Taxa but in the order of the column Order. As I understood it needs to be in factor which is already the case and the data frame is already ordered by the column Order but the output is still in alphabetical order for the column Taxa.
# A tibble: 23 x 4
Order Family Taxa Number
<fct> <fct> <fct> <dbl>
1 Diptere Athericidae Atherix sp. 58
2 Diptere Blephariceridae Blepharicera sp. 10
3 Diptere Blephariceridae Liponeura sp. 23
4 Diptere Chironomidae Chironominae Chironomini 28
5 Diptere Chironomidae Chironominae Tanytarsini 126
6 Diptere Chironomidae Orthocladiinae l.s 184
7 Diptere Empididae Clinocerinae 12
8 Diptere Empididae Hemerodromiinae 13
9 Diptere Limoniidae Dicranota sp. 16
10 Diptere Limoniidae Hexatoma sp. 15
Here's my code and a dput of my data:
ggplot(mydata %>%
arrange(Order, .by_group = T),
aes(x = Taxa, y = Number))
geom_segment(aes(x = Taxa, xend = Taxa, y = 0, yend = Number), color = "grey")
geom_point(size = 3, color = "darkorange")
coord_flip()
structure(list(Order = structure(c(3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 4L, 4L, 4L, 4L, 4L, 4L, 7L, 7L, 7L, 7L, 7L, 7L ), .Label = c("Amphipode", "Coleoptere", "Diptere", "Ephemeroptere", "Plecoptere", "Pulmonata", "Trichoptere"), class = "factor"), Family = structure(c(3L, 5L, 5L, 7L, 7L, 7L, 11L, 11L, 24L, 24L, 33L, 4L, 4L, 4L, 12L, 17L, 17L, 15L, 19L, 19L, 29L, 31L, 31L), .Label = c("Ancylidae", "Anthomyidae", "Athericidae", "Baetidae", "Blephariceridae", "Caenidae", "Chironomidae", "Chloroperlidae", "Dytiscidae", "Elmidae", "Empididae", "Ephemerellidae", "Gammaridae", "Glossomatidae", "Glossosomatidae", "Haliplidae", "Heptageniidae", "Hydraenidae", "Hydropsychidae", "Hydroptilidae", "Leuctridae", "Limnephilidae", "Limniphilidae", "Limoniidae", "Lymnaeidae", "Nemouridae", "Perlidae", "Perlodidae", "Psychomyidae", "Psychomyiidae", "Rhyacophilidae", "Sericostomatidae", "Simuliidae", "Tabanidae", "Tipulidae"), class = "factor"), Taxa = structure(c(3L, 9L, 58L, 13L, 15L, 62L, 19L, 39L, 21L, 42L, 89L, 4L, 5L, 6L, 87L, 29L, 77L, 37L, 23L, 46L, 72L, 51L, 80L), .Label = c("Ancylus fluviatilus", "Antocha sp.", "Atherix sp.", "Baetis alpinus", "Baetis scambus", "Baetis sp.", "Baetis sp. juv", "Baetis vernus", "Blepharicera sp.", "Blephariceridae juv", "Blephariceridae nymphe", "Caenis sp.", "Chironominae Chironomini", "Chironominae Tanypodinae", "Chironominae Tanytarsini", "Chloroperla sp.", "Chloroperlidae", "Chloroperlidae non id", "Clinocerinae", "Clinocerinae nymphe", "Dicranota sp.", "Dinocras sp.", "Diplectrona felix", "Diptere non id", "Diptere nymphe", "Dytiscidae", "Dytiscidae non id juv", "Ecdyonorus sp.", "Ecdyonurus sp.", "Elmis sp.", "Elmis sp. juv", "Epeorus sp.", "Ephemeroptere non id", "Esolus sp.", "Galba truncatula", "Gammarus sp.", "Glossosoma sp.", "Haliplus sp. juv", "Hemerodromiinae", "Hemerodromiinae nymphe", "Heptageniidae non id", "Hexatoma sp.", "Hydracarien", "Hydraena sp.", "Hydroglyphus sp. juv", "Hydropsyche sp.", "Hydropsyche sp. juv", "Hydroptila sp.", "Hydroptilidae non id", "Hyporhyacophila", "Hyporhyacophila sp.", "Leuctra sp.", "Leuctra sp. juv", "Limnephilidae", "Limniphilidae", "Limnophora sp.", "Limoniidae non id", "Liponeura sp.", "Nematode", "Nemura sp.", "Oligochete", "Orthocladiinae l.s", "Pararhyacophila sp.", "Perla maxima", "Perla sp.", "Perlidae non id juv", "Perlodes sp.", "Perlodes sp. juv", "Perlodidae non id juv", "Plecoptere non id", "Protonemura sp.", "Psychomyia pusilla", "Psychomyia sp.", "Psychomyia sp. juv", "Radix", "Rhithrogena semicolorata", "Rhithrogena sp.", "Rhithrogena sp. juv", "Rhyacophila", "Rhyacophila ss", "Rhyacophilidae", "Riolus sp.", "Riolus sp. juv", "Rithrogena sp.", "Scleropracta sp.", "Sericostoma sp.", "Serratella ignita", "Serratella ignita juv", "Simuliidae", "Simuliidae juv", "Simuliidae nymphe", "Siphonoperla sp.", "Tabanidae", "Tipulidae", "Trichoptere non id", "Yola sp. juv" ), class = "factor"), Number = c(58, 10, 23, 28, 126, 184, 12, 13, 16, 15, 302, 309, 4, 108, 140, 36, 148, 28, 38, 386, 7, 5, 26)), row.names = c(NA, -23L), class = c("tbl_df", "tbl", "data.frame"))
I'm sorry for this new question as I think there are many posts already answered but I cannot figure out why it is not working with my data since it's in factor and ordered in the dataframe.
CodePudding user response:
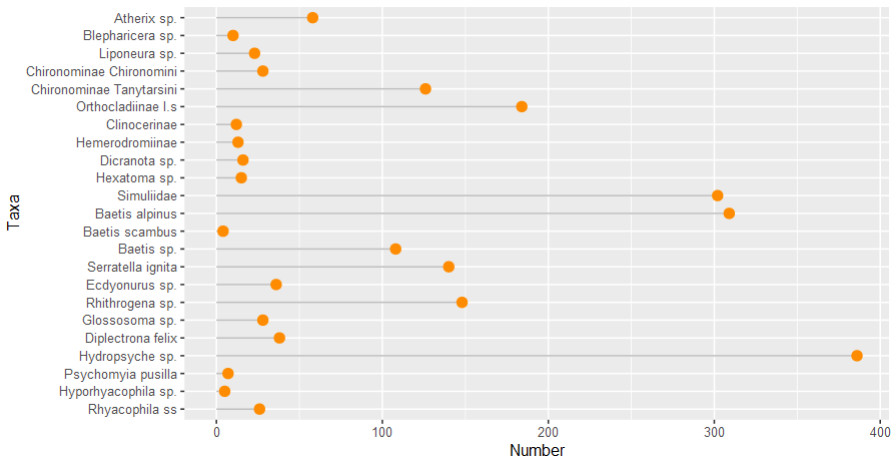
The order of your dataframe actually doesn’t matter — what matters is the order of the factor levels. You can order factor levels based on another variable using forcats::fct_reorder():
library(tidyverse)
# use as.integer to order by underlying factor codes
mydata %>%
mutate(Taxa = fct_reorder(Taxa, as.integer(Order))) %>%
ggplot(aes(x = Taxa, y = Number))
geom_segment(aes(x = Taxa, xend = Taxa, y = 0, yend = Number), color = "grey")
geom_point(size = 3, color = "darkorange")
coord_flip()
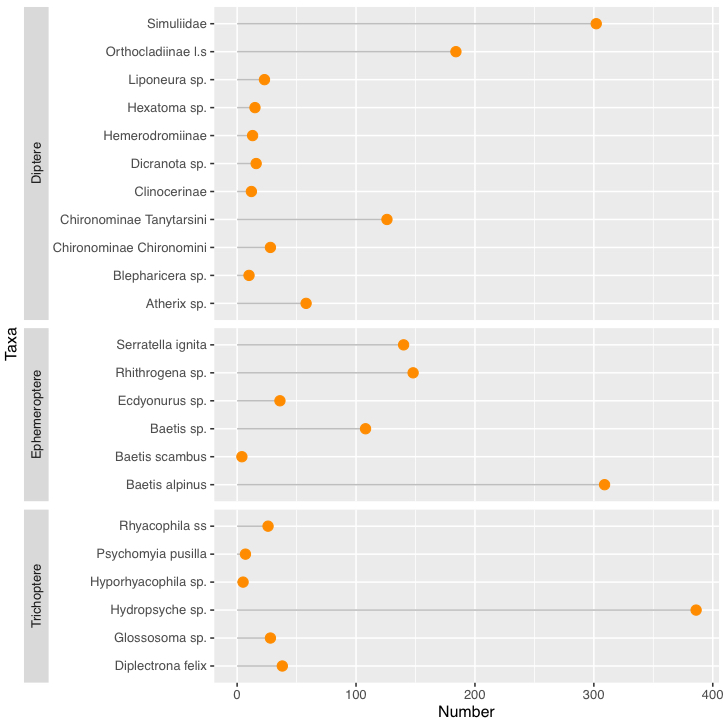
Alternatively, you could facet by Order:
ggplot(mydata, aes(x = Taxa, y = Number))
geom_segment(aes(x = Taxa, xend = Taxa, y = 0, yend = Number), color = "grey")
geom_point(size = 3, color = "darkorange")
coord_flip()
facet_grid(Order ~ ., scales = "free_y", space = "free_y", switch = "y")
theme(strip.placement = "outside")
CodePudding user response:
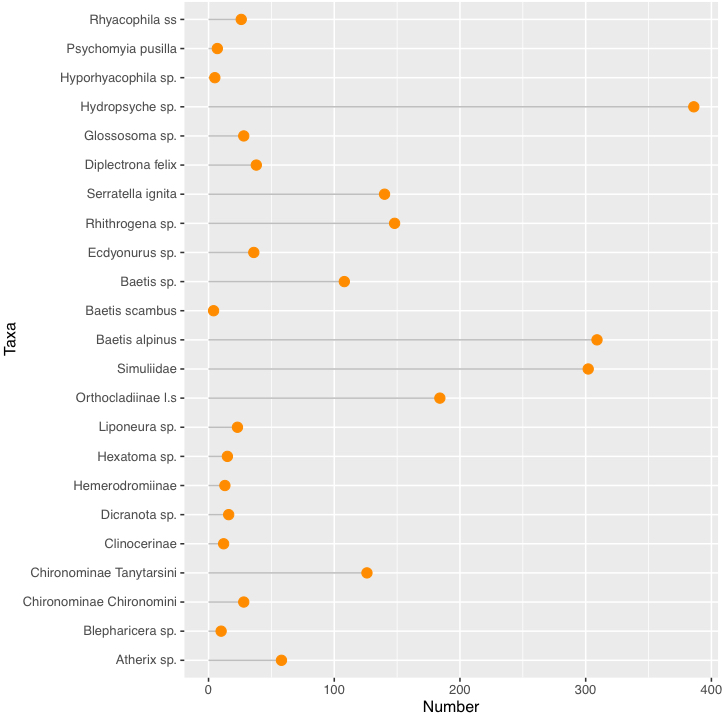
If you want to make the factor appears as in the data, refactor the variable with levels as unique(your_column), which will use the levels as they appear in the dataset.
Your points are ordered in the reversed order, just use scale_x_discrete(limits=rev) to make it appear the correct way:
mydata %>%
mutate(Taxa = factor(Taxa, levels = unique(Taxa))) %>%
ggplot(aes(x = Taxa, y = Number))
geom_segment(aes(x = Taxa, xend = Taxa, y = 0, yend = Number), color = "grey")
geom_point(size = 3, color = "darkorange")
coord_flip()
scale_x_discrete(limits=rev)
More thorough explanation:
Factor is a class and contains levels. ggplot orders the factor variable in the plot according to the order of the levels (so not according to the order as they appear in the data). If you order the dataset, the order of the levels will remain the same, i.e., you have to explicitly reorder the levels to do that. So one way to reorder the levels is to use factor and explicitly set the levels in the order of the data using unique. It may be a bit cumbersome, but that's the cleanest way to deal with factors.