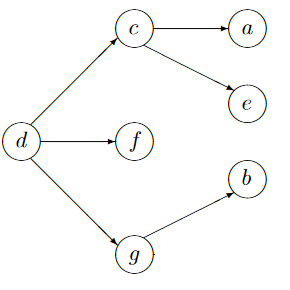
I have a graph g
n = 8
m <- t(matrix(c(
0,0,0,0,0,0,0,8,
3,0,0,0,0,0,0,0,
5,0,0,5,1,0,0,0,
0,0,6,0,0,7,1,0,
0,6,2,0,0,0,0,0,
0,0,0,0,0,0,0,0,
7,4,0,0,8,0,0,3,
0,3,0,0,0,9,0,0),ncol=n))
library(igraph)
g <- graph_from_adjacency_matrix(m, weighted=TRUE, mode="directed")
V(g1)$name <- letters[1:n]
After some calculation I have the vector str with vertex's names:
str <- c("dca", "dgb", "dc", NA, "dce", "df", "dg", "dg")
I need to find the unique nodes and plot a tree where root node is the first symbol in str.
My attempt is:
str[is.na(str)]<-"";
vname <- noquote(unique(strsplit((paste0(str, collapse="")),"")[[1]]))
vname
# d c a g b e f
vid <- match( vname, V(g1)$name )
vid
# 4 3 1 7 2 5 6
One can see that the difference between nodes of g and vid is the h node, its id is 8.
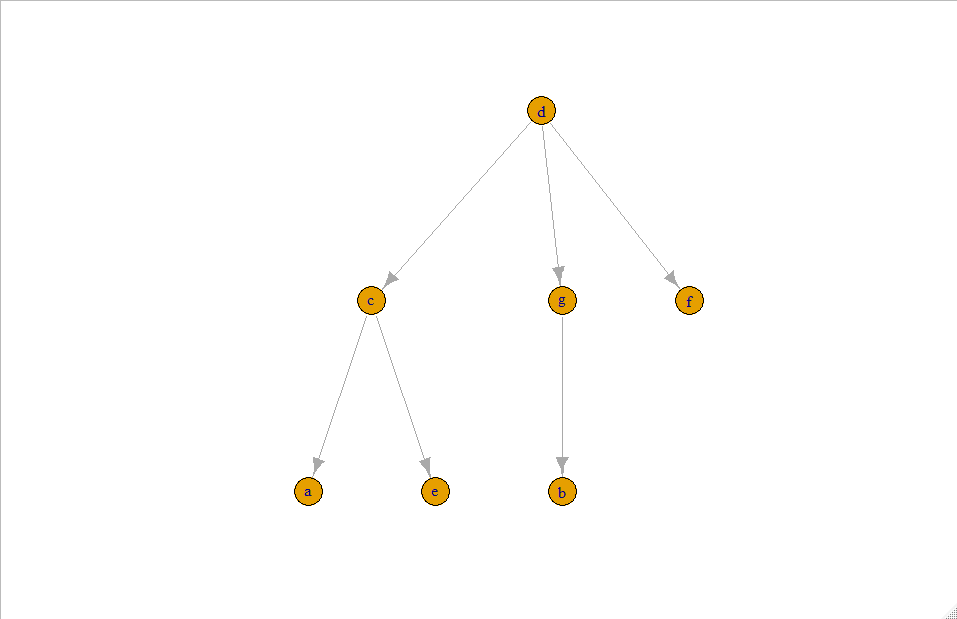
Expected result is:
Question. How to define the difference in id's nodes and delete vertices with corresponding edges to plot a tree?
CodePudding user response:
You can use subgraph.edges:
# find wanted edges
str <- unique(str[!is.na(str)])
keep_edges <- apply(
X = unique(
do.call(rbind, lapply(strsplit(str, ""), function(x) embed(x, 2)[,2:1]))
),
MARGIN = 1,
paste0,
collapse = "|"
)
# subgraph containing only wanted edges
g <- subgraph.edges(g1, keep_edges)
nodes <- V(g)$name
plot(g, layout = igraph::layout_as_tree)
CodePudding user response:
You can use eval graph_from_literal gsub to create a graph object g like below
g <- eval(
str2lang(
sprintf(
"graph_from_literal(%s)",
toString(gsub("(?<=.)(?=.)", "- ", na.omit(str), perl = TRUE))
)
)
)
and you will see
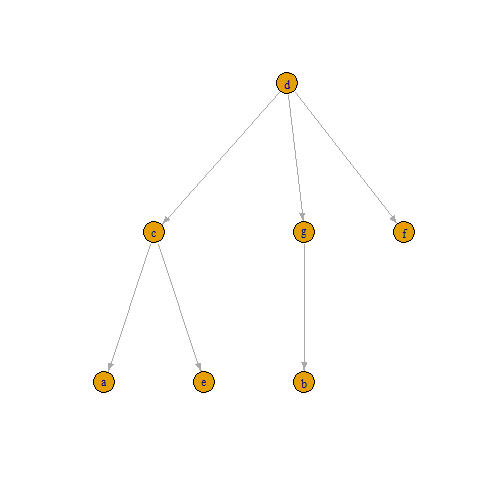
> g
IGRAPH fe5b7ea DN-- 7 6 --
attr: name (v/c)
edges from fe5b7ea (vertex names):
[1] d->c d->g d->f c->a c->e g->b
and plot(g, layout = layout_as_tree) gives