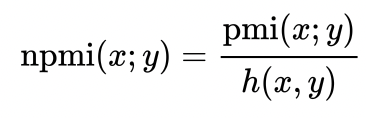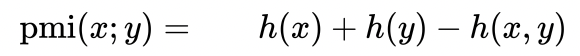I have a matrix dat defined as follows:
dat <- matrix(c(0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1,
0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 0, 0, 1,
2, 3, 1, 1, 3, 2, 1, 3, 2, 2, 1, 3, 3, 2, 1, 2, 1, 2, 2, 2,
1, 2, 1, 1, 2, 2, 2, 2, 2, 2, 1, 2, 1, 1, 1, 2, 2, 1, 1, 1), ncol = 4)
I can calculate the numerator as follows:
apply(combn(1:ncol(dat), 2), 2, function(i) mutual_info(dat[, i], local = TRUE))
How can i find the denominator in order two divide the above and find the mpmi?
CodePudding user response:
Assuming that h is the entropy and mutual_info is the same function as widyr::pairwise_pmi you can do this:
library(tidyverse)
library(widyr)
library(DescTools)
dat <- matrix(c(
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1,
0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 0, 0, 1,
2, 3, 1, 1, 3, 2, 1, 3, 2, 2, 1, 3, 3, 2, 1, 2, 1, 2, 2, 2,
1, 2, 1, 1, 2, 2, 2, 2, 2, 2, 1, 2, 1, 1, 1, 2, 2, 1, 1, 1
), ncol = 4)
dat <- as_tibble(dat)
#> Warning: The `x` argument of `as_tibble.matrix()` must have unique column names if `.name_repair` is omitted as of tibble 2.0.0.
#> Using compatibility `.name_repair`.
#> This warning is displayed once every 8 hours.
#> Call `lifecycle::last_lifecycle_warnings()` to see where this warning was generated.
dat %>%
colnames() %>%
combn(2) %>%
t() %>%
as_tibble() %>%
mutate(
pmi = list(V1, V2) %>% pmap(~ pairwise_pmi(tibble(from = dat[[.x]], to = dat[[.y]]), from, to))
) %>%
unnest() %>%
distinct(V1, V2, pmi) %>%
mutate(
h = list(V1, V2, pmi) %>% pmap_dbl(~ Entropy(dat[[.x]], dat[[.y]])),
npmi = pmi / h
)
#> Warning: `cols` is now required when using unnest().
#> Please use `cols = c(pmi)`
#> # A tibble: 6 × 5
#> V1 V2 pmi h npmi
#> <chr> <chr> <dbl> <dbl> <dbl>
#> 1 V1 V2 0 1.32 0
#> 2 V1 V3 -0.0770 2.15 -0.0358
#> 3 V1 V4 0 1.68 0
#> 4 V2 V3 0 2.33 0
#> 5 V2 V4 0 1.80 0
#> 6 V3 V4 0 2.44 0
Created on 2022-04-06 by the reprex package (v2.0.0)
e.g. where V3 is the third column of the matrix.