I have a graph similar to the following:
library(plotly)
df <- as.data.frame(1:19)
df$CATEGORY <- c("C","C","A","A","A","B","B","A","B","B","A","C","B","B","A","B","C","B","B")
df$x <- c(126,40,12,42,17,150,54,35,21,71,52,115,52,40,22,73,98,35,196)
df$y <- c(92,62,4,23,60,60,49,41,50,76,52,24,9,78,71,25,21,22,25)
df[,1] <- NULL
df$fv <- df %>%
filter(!is.na(x)) %>%
lm(y ~ x*CATEGORY,.) %>%
fitted.values()
p <- plot_ly(data = df,
x = ~x,
y = ~y,
color = ~CATEGORY,
type = "scatter",
mode = "markers"
) %>%
add_trace(x = ~x, y = ~fv, mode = "lines")
p
It works fine since I need to have multiple regression line on the same plot, but what I would really need is polynomial regression lines for each category. I tried to replace "lm(y ~ x*CATEGORY,.) " with the following:
df1$fv <- df1 %>%
filter(!is.na(x)) %>%
lm(y ~ poly(x*CATEGORY,.),2) %>%
fitted.values()
but it doesn't work. Any suggestions? Thank you
CodePudding user response:
@Dave2e is correct about the mis-specification. To generate a plot that looks good (i.e., one that generates a polynomial-looking curve, you might need to do a bit more work:
library(plotly)
library(ggeffects)
library(dplyr)
library(tidyr)
df <- as.data.frame(1:19)
df$CATEGORY <- c("C","C","A","A","A","B","B","A","B","B","A","C","B","B","A","B","C","B","B")
df$x <- c(126,40,12,42,17,150,54,35,21,71,52,115,52,40,22,73,98,35,196)
df$y <- c(92,62,4,23,60,60,49,41,50,76,52,24,9,78,71,25,21,22,25)
df[,1] <- NULL
df <- df %>% arrange(CATEGORY, x)
hyp <- by(df$x, list(df$CATEGORY), function(x)seq(min(x), max(x), length=50))
hyp <- do.call(data.frame, hyp) %>%
pivot_longer(everything(), names_to = "CATEGORY", values_to="x")
mod <- lm(y ~ poly(x, 2)*CATEGORY, data=df)
hyp$predicted <- predict(mod, newdata=hyp)
p <- plot_ly() %>%
add_trace(data = df,
x = ~x,
y = ~y,
color = ~CATEGORY,
type = "scatter",
mode = "markers") %>%
add_trace(data = hyp, x = ~x, y = ~predicted, color = ~CATEGORY, mode = "lines")
p
#> No trace type specified:
#> Based on info supplied, a 'scatter' trace seems appropriate.
#> Read more about this trace type -> https://plotly.com/r/reference/#scatter
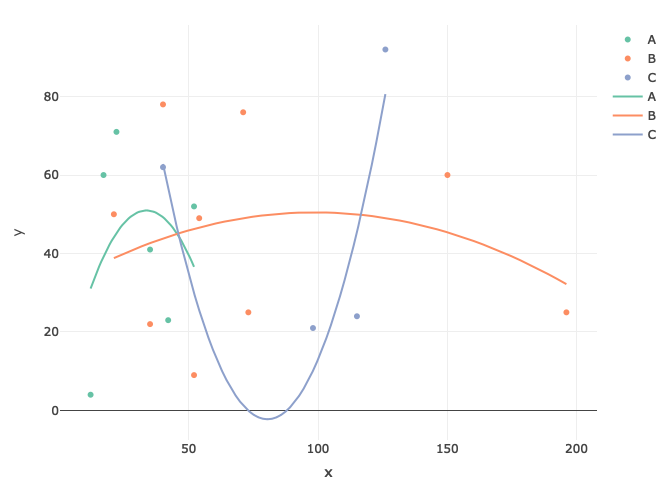
Created on 2022-04-07 by the reprex package (v2.0.1)
CodePudding user response:
Your formula is incorrect. Try:
df %>%
filter(!is.na(x)) %>%
lm(y ~ poly(x,2, raw=TRUE)*CATEGORY, data=.) %>%
fitted.values()
- df1 is not defined in your sample code, so assuming df here.
- Use the data=. to reference the data source.
- Inside the poly function define the power, in this case 2.
- Move CATEGORY outside the poly function. x* CATEGORY x^2* CATEGORY etc.
