I have the models with overdispersion and almost 40% of the data set are true zeros with biological meaning. I'd like to study the temperature (temp) and time storage in days (storage) influence on the insect's development (development), but the storage is temporally correlated. This is a classic zero dilemma case, inflated or not inflated models?
I try to do:
#Packages
library(lme4)
library(glmmTMB)
library(tidyverse)
library(bbmle)
library(broom)
#Open my dataset
myds<-read.csv("https://raw.githubusercontent.com/Leprechault/trash/main/ds.desenvol.csv")
str(myds)
# 'data.frame': 400 obs. of 4 variables:
# $ temp : num 0 0 0 0 0 0 0 0 0 0 ...
# $ storage : int 5 5 5 5 5 5 5 5 5 5 ...
# $ rep : chr "r1" "r2" "r3" "r4" ...
# $ development: int 0 23 22 27 24 25 24 22 0 22 ...
# Zero proportion - All true zeros with biological meaning
p.0 <- myds[myds$development==0,]
(length(p.0[,1])/length(myds[,1]))*100
# [1] 39.25 % of the data set are zeros
# Models creation Negative binomial
# 1 - Model not inflated
m_1 <- glmmTMB(development ~ temp storage (1 | storage), data = myds,
family = nbinom2, zi = ~ 0)
# 3 - Model inflated
m_2 <- glmmTMB(development ~ temp storage (1 | storage), data = myds,
family = nbinom2, zi = ~ 1)
# Put it all the models together:
modList <- tibble::lst(m_1,m_2)
bbmle::AICtab(modList)
# dAIC df
# m_2 0 6
# m_1 929 5
#
But, is difficult for me, the adoption of highly complex models with just AIC metric as support. Please, any help other support decision criteria for model selection in this case?
CodePudding user response:
There's a bunch going on here.
First let's look at the data:
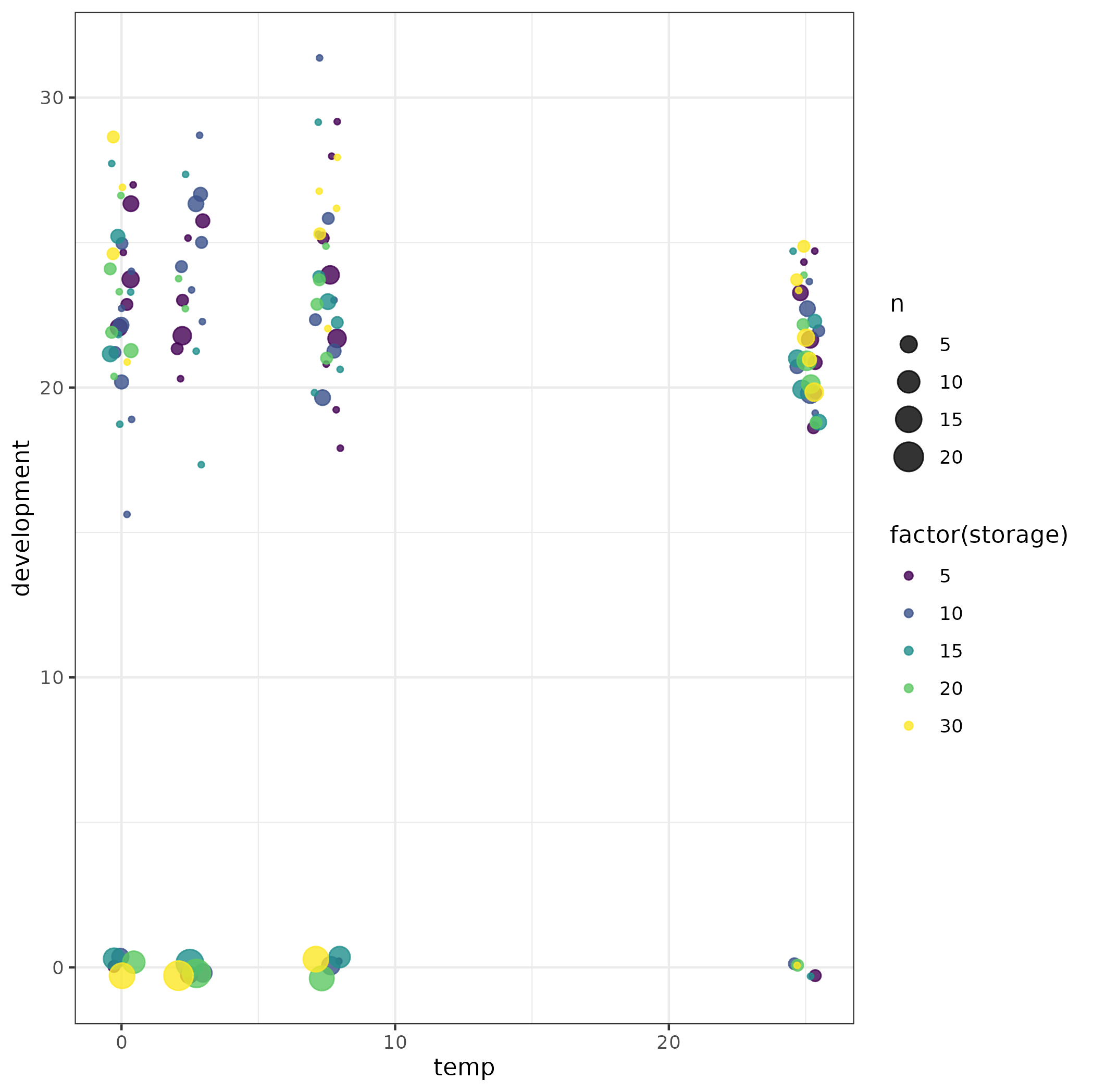
library(ggplot2); theme_set(theme_bw())
(ggplot(myds)
aes(temp, development, colour = factor(storage))
stat_sum(position = position_jitter(width = 0.5),
alpha = 0.8)
scale_color_viridis_d()
)
Just from looking at the picture, it's pretty obvious that a zero-inflated model is necessary (there's no way a non-zero-inflated Poisson or negative binomial can produce a bimodal distribution with many values at zero and many values far from zero).
Fit first model:
m_1 <- glmmTMB(development ~ temp storage (1 | storage), data = myds,
family = nbinom2)
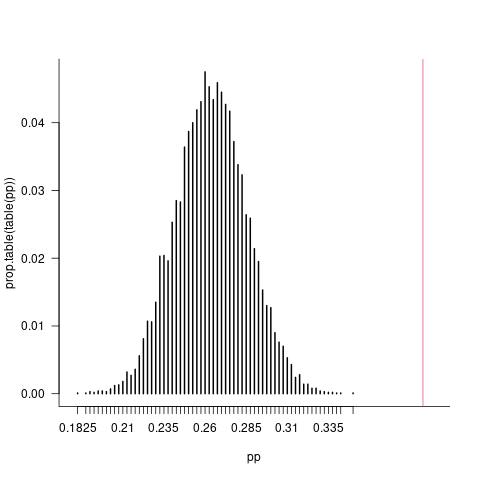
Simulate the model many times to look at the expected distribution of proportion zero values:
set.seed(101)
system.time(
pp <- replicate(10000, mean(simulate(m_1)[[1]] == 0))
)
par(las=1, bty = "l")
plot(prop.table(table(pp)), xlim = c(0.18, 0.40))
abline(v = p.0, col = 2)
(10,000 samples is overkill here.) Again, obvious that there are way too many zero values in the response to be explained by the best-fitting NB model (p < 0.0001, since the observed proportion [0.39, red line] is well above the maximum proportion in 10,000 replicates (0.35).
Let's dig in farther.
When I fit a model with zero-inflation I get model convergence warnings (this will vary across platforms), and a huge value of sigma(m_2):
m_2 <- update(m_1, ziformula = ~ 1)
## Warning messages:
## 1: In fitTMB(TMBStruc) :
Model convergence problem; non-positive-definite Hessian matrix. See vignette('troubleshooting')
## 2: In fitTMB(TMBStruc) :
Model convergence problem; false convergence (8). See vignette('troubleshooting')
sigma(m_2)
## [1] 262965681398
This means that a negative binomial model isn't appropriate, and we should revert to a Poisson response
m_3 <- update(m_1, ziformula = ~ 1, family = poisson)
We can see that the estimated random effect is very small (effectively zero):
VarCorr(m_3)
Conditional model:
Groups Name Std.Dev.
storage (Intercept) 8.9098e-06
This means that the Poisson variation is enough to explain any deviations around a linear effect of storage, we don't need the random effect (although it doesn't really hurt to keep it in).
m_4 <- update(m_1, ziformula = ~ 1, family = poisson,
. ~ . - (1|storage))
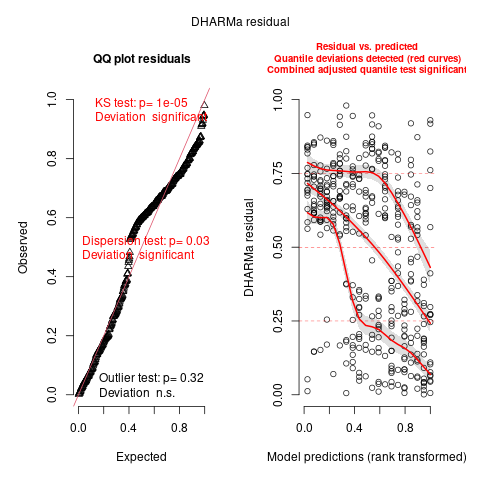
However, checking residuals with DHARMa, we do still have a bit of a problem:
library(DHARMa)
plot(s4 <- simulateResiduals(m_4))
It's often easiest to understand what's going on by plotting residuals vs individual factors:
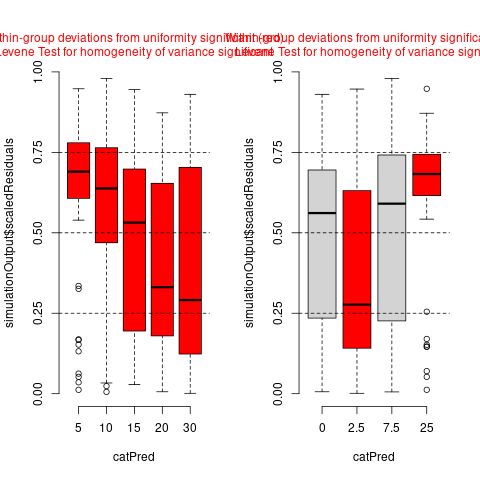
par(mfrow=c(1,2))
plotResiduals(s4, myds$storage, rank = FALSE)
plotResiduals(s4 myds$temp, rank = FALSE)
This shows fairly severe heteroscedasticity: the storage == 5 and temp == 25 conditions are much less variable than the other categories (storage == 10 is somewhat less variable).
Unfortunately, it's hard to incorporate heteroscedasticity in a Poisson model (because the variance is completely determined once we know the mean). I tried a negative binomial model without the random effect and with heteroscedasticity by storage and temp (which didn't work very well), then tried a hurdle (zero-inflated) log-link Gamma with heteroscedasticity, which so far is the best of the bunch:
m_5 <- update(m_2,
. ~ . - (1|storage),
dispformula = ~storage temp)
m_6 <- update(m_2,
family = ziGamma(link = "log"),
. ~ . - (1|storage),
dispformula = ~storage temp)
# Put all the models together:
modList <- list(nb2=m_1,
nb2_zi=m_2,
pois_zi=m_3,
pois_zi_noRE = m_4,
nb2_zi_het = m_5,
gamma_zi_het = m_6)
bbmle::AICtab(modList)
dAIC df
gamma_zi_het 0.0 7
pois_zi_noRE 197.3 4
pois_zi 199.3 5
nb2 1130.3 5
nb2_zi NA 6
nb2_zi_het NA 7
The DHARMa output still doesn't look great, but it's a lot better than when we started ...