To visualize the results of my linear mixed model (LMM) I would like to plot spagetti plots that track the change in volume1 over time for all participants. I would like to combine this with plotting the intercept and coefficient (including 95% confidence intervals) of my LMM as an overlay. However, so far I have not succeeded to combine these plots and I am looking for some help.
- For my LMM I used the 'lmer' function that is part of the lme4 package in R.
Here is the first part of my code to run the linear mixed model and obtain spagetti plots for all measurments over time per subject
#Load subset of my data that I obtained with the 'dput' function
Longitudinal_measures_volume <- structure(list(id = structure(c(1L, 2L,
3L, 9L, 10L, 11L, 13L, 18L, 19L, 1L, 2L, 3L, 9L, 10L, 11L, 13L, 18L, 3L, 9L, 11L, 13L, 18L, 19L), .Label = c("01_01", "01_02", "01_05", "01_06", "01_07", "01_08", "01_11", "01_12", "01_13", "01_14", "01_17", "01_19", "01_21", "01_22", "01_24", "01_25", "01_27", "01_28", "01_30", "01_33", "01_35", "02_01", "02_02","02_03", "02_04", "02_05", "02_06", "02_07", "02_09", "02_10","02_11", "02_12", "02_13", "02_14", "02_15", "02_18", "02_20", "02_21", "02_23", "02_24", "02_25", "03_01", "03_02", "03_03", "03_04", "03_05", "03_06", "03_07", "03_08", "03_09", "03_10","03_11", "03_12", "03_13", "03_14", "03_15", "03_16", "03_18","03_19", "03_20"), class = "factor"), Time = c(67, 41, 41, 46, 39, 42, 38, 41, 47, 119, 105, 111, 95, 95, 98, 103, 95, 195, 193, 196, 222, 188, 211), Volume1 = c(3.45897015360706, 0.898545274090074, -1.46885896648035, 0.48984364347097, 2.52266602788419, 0.930927639876306, -1.79340703526533, 2.58022117920645, -1.88239745399949,3.22242849165164, 0.615590786285958, -1.14628038654842, 0.996278646866944, 2.16556421777876, 1.64058648629247, -1.13969644900845, 2.45418152346369, -0.307093266274559, 1.51877037424232, 1.74322136278721, -1.72984060264199, 1.96975730534075, -1.25357412221767)), row.names = c(1L, 2L, 3L, 8L, 9L, 10L, 12L, 17L, 18L, 60L, 61L, 62L, 68L, 69L, 70L, 72L, 77L, 79L, 85L, 86L, 88L, 93L, 94L), class = "data.frame")
#load packages
library(lme4)
library(ggeffects)
#LMM model (example)
LMMmodel_example <- lmer(Volume1 ~ Time (1 |id), data=Longitudinal_measures_volume)
summary(LMMmodel_example)
#Obtain spagetti plots for all measurements over time per subject
plot <- ggplot(data=Longitudinal_measures_volume, aes(x=Time, y=Volume1, group =id)) geom_line(aes(x=Time, y=Volume1, color=id))
print(plot)
Then I wrote the following, separate, line of code to plot the intercept and coefficient(with 95% confidence interval) of my LMM:
Group_line <- ggpredict(LMMmodel_example, terms = c("Time"), interval = "confidence", ci.lvl=0.95)
plot(Group_line)
Now, I would want to combine these two plots into one, where I have the spagetti plots and a plot with the intercept and coefficient (including 95% confidence interval) of my LMM as an overlay. My first attempt was the following:
plot_overlay <- ggplot(data=Longitudinal_measures_volume, aes(x=Time, y=Volume1, group =id)) geom_line(aes(x=Time, y=Volume1, color=id)) ggpredict(LMMmodel_example, terms = c("Time"), interval = "confidence", ci.lvl=0.95)
I then received the following error statement:
Error in ggplot(data = Longitudinal_measures_volume, aes(x = Time, y = Volume1, :
non-numeric argument to binary operator
In addition: Warning message:
Incompatible methods (" .gg", "Ops.data.frame") for " "*
I thought that the error might be due to that the 'ggpredict' function is now looking for the same aesthetics as specfied with 'ggplot'. As a potential solution I tried to add 'inherit.aes=FALSE' to the ggpredict function, but unfortunately this didn't solve my error.
I would be grateful for any help to combine these plots, thanks a lot in advance! Recommendations for other functions are also very welcome.
CodePudding user response:
Is it possible to have a reproducible example please? the model you provide does not work in my R so I don't get the problem on the ggplot.
Maybe you can think about the geom_ribbon function, with something like :
ggplot(data=Longitudinal_measures_volume, aes(x=Time, y=Volume1, group =id)) geom_line(aes(x=Time, y=Volume1, color=id))
geom_ribbon(aes(ymin = minpred, ymax = maxpred), alpha = 0.1)
CodePudding user response:
The output of ggeffects can be converted to a data frame, which can be used to add a geom_ribbon and geom_line to your existing plot.
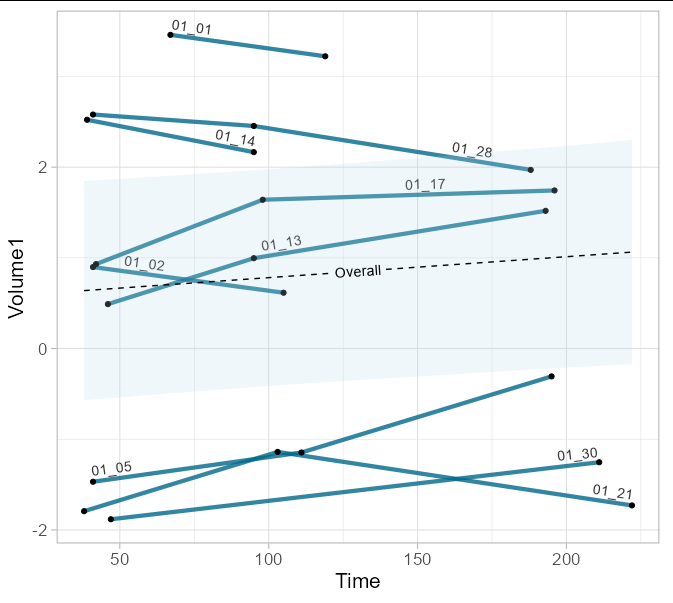
I think there are too many colors to make your individual ID values identifiable on the plot, so I have created a version with direct labelling as an option:
library(geomtextpath)
plot <- ggplot(data=Longitudinal_measures_volume,
aes(x = Time, y = Volume1, group = id))
geom_textline(aes(x = Time, y = Volume1, label = id, hjust = id),
vjust = -0.3,
linewidth = 1.5, alpha = 0.8,
linecolor = 'deepskyblue4')
geom_point()
scale_hjust_manual(values = c(0, 0.2, 0, 0.4, 1, 0.75, 1, 0.9, 1))
theme_light(base_size = 16)
Group_line <- ggpredict(LMMmodel_example, terms = c("Time"),
interval = "confidence", ci.lvl = 0.95)
plot
geom_ribbon(aes(x = x, ymax = conf.high, ymin = conf.low),
data = as.data.frame(Group_line), inherit.aes = FALSE,
alpha = 0.2, fill = 'lightblue')
geom_textline(aes(x = x, y = predicted, label = 'Overall'), linetype = 2,
data = as.data.frame(Group_line), inherit.aes = FALSE)