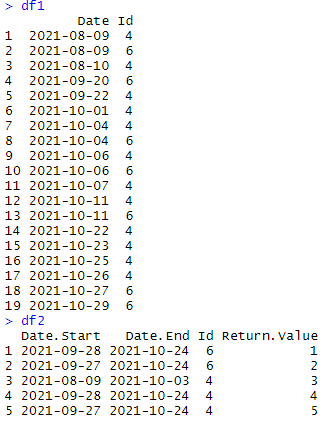
I have 2 dataframes, one with list of dates (df1) and another one with date ranges by ID (df2). I would like to add a column from df2 if a date in df1 sits within the date range in df2.
## This is my base data
df1 <-
structure(list(Date = structure(c(18848, 18848, 18849, 18890,
18892, 18901, 18904, 18904, 18906, 18906, 18907, 18911, 18911,
18922, 18923, 18925, 18926, 18927, 18929), class = "Date"), Id = c(4,
6, 4, 6, 4, 4, 4, 6, 4, 6, 4, 4, 6, 4, 4, 4, 4, 6, 6)), row.names = c(NA,
19L), class = "data.frame")
## This is my date range table
df2 <- structure(list(Date.Start = structure(c(18898, 18897, 18848,
18898, 18897), class = "Date"), Date.End = structure(c(18924,
18924, 18903, 18924, 18924), class = "Date"), Id = c(6, 6, 4,
4, 4), Return.Value = c(1, 2, 3, 4, 5)), row.names = c(NA, 5L
), class = "data.frame")
So for the first row of df1, Date = '2021-08-09' sits within date range '2021-08-09' to '2021-10-03' (row 3 of df2) so I want to add a Return Value column to df1 with a value of 3.
There is no match for row 2 in df1 so this will return nothing.
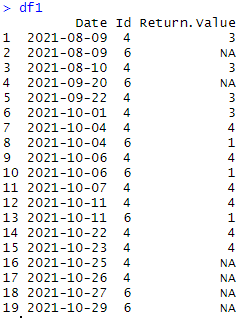
My expected output is:
I tried this link Check if a date is in range of lookup table but I want to add a column from df2 instead of returning logical vector.
CodePudding user response:
data.table version using the mult="first" option on a non-equi join allowing >= and <= comparisons:
library(data.table)
setDT(df1)
setDT(df2)
df1[, Return.Value := df2[
df1, on=c("Id", "Date.Start<=Date", "Date.End>=Date"), Return.Value, mult="first"]]
df1
# Date Id Return.Value
# 1: 2021-08-09 4 3
# 2: 2021-08-09 6 NA
# 3: 2021-08-10 4 3
# 4: 2021-09-20 6 NA
# 5: 2021-09-22 4 3
# 6: 2021-10-01 4 3
# 7: 2021-10-04 4 4
# 8: 2021-10-04 6 1
# 9: 2021-10-06 4 4
#10: 2021-10-06 6 1
#11: 2021-10-07 4 4
#12: 2021-10-11 4 4
#13: 2021-10-11 6 1
#14: 2021-10-22 4 4
#15: 2021-10-23 4 4
#16: 2021-10-25 4 NA
#17: 2021-10-26 4 NA
#18: 2021-10-27 6 NA
#19: 2021-10-29 6 NA
CodePudding user response:
It's pretty messy...
library(dplyr)
df1 %>%
left_join(df2, by = 'Id') %>%
rowwise %>%
mutate(Return.Value = ifelse(between(Date, Date.Start, Date.End), Return.Value, NA)) %>%
select(-Date.Start, -Date.End) %>%
group_by(Date, Id) %>%
filter({if (sum(!is.na(Return.Value) >0)) !is.na(Return.Value) else row_number() == 1}) %>%
filter(row_number() == 1)
Date Id Return.Value
<date> <dbl> <dbl>
1 2021-08-09 4 3
2 2021-08-09 6 NA
3 2021-08-10 4 3
4 2021-09-20 6 NA
5 2021-09-22 4 3
6 2021-10-01 4 3
7 2021-10-04 4 4
8 2021-10-04 6 1
9 2021-10-06 4 4
10 2021-10-06 6 1
11 2021-10-07 4 4
12 2021-10-11 4 4
13 2021-10-11 6 1
14 2021-10-22 4 4
15 2021-10-23 4 4
16 2021-10-25 4 NA
17 2021-10-26 4 NA
18 2021-10-27 6 NA
19 2021-10-29 6 NA
CodePudding user response:
We could do this also with match_fun() function from fuzzyjoin package:
library(dplyr)
library(fuzzyjoin)
fuzzy_left_join(
df1, df2,
by = c(
"Id" = "Id",
"Date" = "Date.Start",
"Date" = "Date.End"
),
match_fun = list(`==`, `>=`, `<=`)) %>%
group_by(Date, Id.x) %>%
slice(1) %>%
dplyr::select(Date, Id = Id.x, Return.Value)
Date Id Return.Value
<date> <dbl> <dbl>
1 2021-08-09 4 3
2 2021-08-09 6 NA
3 2021-08-10 4 3
4 2021-09-20 6 NA
5 2021-09-22 4 3
6 2021-10-01 4 3
7 2021-10-04 4 4
8 2021-10-04 6 1
9 2021-10-06 4 4
10 2021-10-06 6 1
11 2021-10-07 4 4
12 2021-10-11 4 4
13 2021-10-11 6 1
14 2021-10-22 4 4
15 2021-10-23 4 4
16 2021-10-25 4 NA
17 2021-10-26 4 NA
18 2021-10-27 6 NA
19 2021-10-29 6 NA