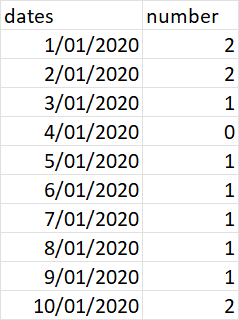
I have 2 tables
df1 = data.frame("dates" = c(seq(as.Date("2020-1-1"), as.Date("2020-1-10"), by = "days")))
df2 = data.frame("observations" = c("a", "b", "c", "d"), "start" = as.Date(c("2019-12-30", "2020-1-1", "2020-1-5","2020-1-10")), "end"=as.Date(c("2020-1-3", "2020-1-2", "2020-1-12","2020-1-14")))
I would like to know the number of observation periods that occur on each day of df1, based on the start/stop dates in df2. E.g. on 1/1/2020, observations a and b were in progress, hence "2".
The expected output would be as follows:
I've tried using sums
df1$number = sum(as.Date(df2$start) <= df1$dates & as.Date(df2$end)>=df1$dates)
But that only sums up the entire column values
I've then tried to create a custom function for this:
df1$number = apply(df1, 1, function(x) sum(df2$start <= x & df2$end>=x))
But it returns an NA value.
I then tried to do embed an "ifelse" within it, but get the same issue with NAs
apply(df1, 1, function(x) sum(ifelse(df2$start <= x & df2$end>=x, 1, 0)))
Can anyone suggest what the issue is? Thanks!
edit: an interval join was suggested which is not what I'm trying to get - I think naming the observations with a numeric label was what caused confusion. I am trying to find out the TOTAL number of observations with periods that fall within the day, as compared to doing a 1:1 match.
Regards Sing
CodePudding user response:
Here is a potential solution using dplyr/tidyverse functions and the %within% function from the lubridate package. This approach is similar to Left Join Subset of Column Based on Date Interval, however there are some important differences i.e. use summarise() instead of filter() to avoid 'losing' dates where "number" == 0, and join by 'character()' as there are no common columns between datasets:
library(dplyr)
library(lubridate)
df1 = data.frame("dates" = c(seq(as.Date("2020-1-1"),
as.Date("2020-1-10"),
by = "days")))
df2 = data.frame("observations" = c("1", "2", "3", "4"),
"start" = as.Date(c("2019-12-30", "2020-1-1", "2020-1-5","2020-1-10")),
"end"=as.Date(c("2020-1-3", "2020-1-2", "2020-1-12","2020-1-14")))
df1 %>%
full_join(df2, by = character()) %>%
mutate(number = dates %within% interval(start, end)) %>%
group_by(dates) %>%
summarise(number = sum(number))
#> # A tibble: 10 × 2
#> dates number
#> <date> <dbl>
#> 1 2020-01-01 2
#> 2 2020-01-02 2
#> 3 2020-01-03 1
#> 4 2020-01-04 0
#> 5 2020-01-05 1
#> 6 2020-01-06 1
#> 7 2020-01-07 1
#> 8 2020-01-08 1
#> 9 2020-01-09 1
#> 10 2020-01-10 2
Created on 2022-06-27 by the reprex package (v2.0.1)
Does this approach work with your actual data?
CodePudding user response:
Define the comparison in a function f and pass it through outer, rowSums is what you're looking for.
f <- \(x, y) df1[x, 1] >= df2[y, 2] & df1[x, 1] <= df2[y, 3]
cbind(df1, number=rowSums(outer(1:nrow(df1), 1:nrow(df2), f)))
# dates number
# 1 2020-01-01 2
# 2 2020-01-02 2
# 3 2020-01-03 1
# 4 2020-01-04 0
# 5 2020-01-05 1
# 6 2020-01-06 1
# 7 2020-01-07 1
# 8 2020-01-08 1
# 9 2020-01-09 1
# 10 2020-01-10 2