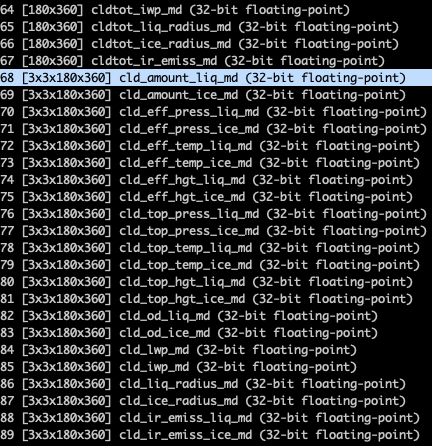
I'd like to plot an array with the following dimensions: [3x3x180x360]
it's a 180x360 world map with 3 different opacity layers and 3 different pressure levels. Thus, I would like to plot the map with data of 1 opacity layer at one pressure level.
How can I access the array and plot the data using plt.imshow() for example?
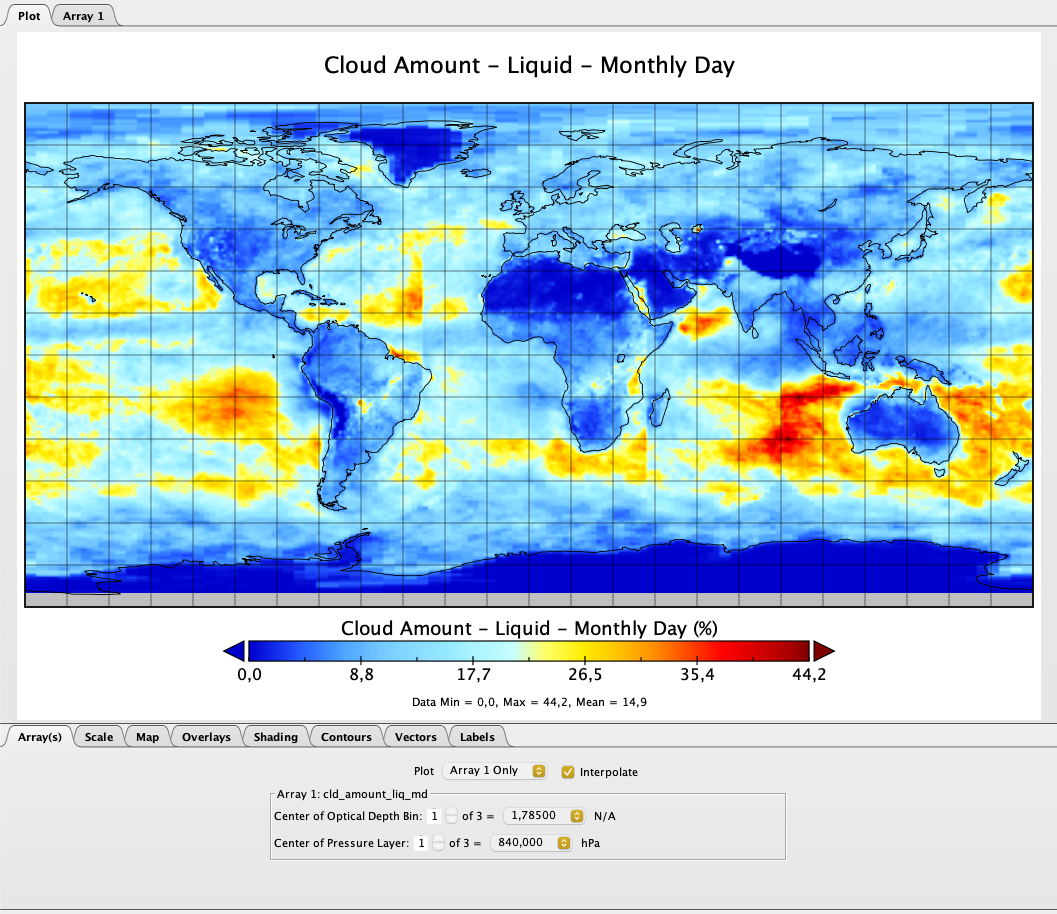
Visualizing the data with panoply looks the following way:
import numpy as np
from osgeo import gdal
granule = "CER_CldTypHist_GEO-MODIS_Edition4A_407408.202109.hdf"
hdf_file = gdal.Open(workdir_data "/" granule)
subDatasets = hdf_file.GetSubDatasets()
cld_amount_liq_md = gdal.Open(subDatasets[68][0]).ReadAsArray() # takes ~5min to read ...
# Shape of cld_amount_liq_md after ReadAsArray() is [64800, 3, 3] hence:
cld_amount_liq_md = cld_amount_liq_md.reshape(180,360, 3, 3)
#filtering bad data:
cld_amount_liq_md[cld_amount_liq_md > 3.40E38] = np.nan
# Plotting -> how can I plot the data?
plt.imshow(cld_amount_liq_md[?,?,?] ,cmap ="jet")```
Update: Here's additional info due to inputs from the comments:
- How do I get the dimensions from the HDF file?
from osgeo import gdal
granule = "CER_CldTypHist_GEO-MODIS_Edition4A_407408.202109.hdf"
hdf_file = gdal.Open(workdir_data "/" granule)
subDatasets = hdf_file.GetSubDatasets()
j=0
for i in subDatasets:
print(j, i[1]) # i[0] contains the path to subdataset, i[1] the info as seen in the screenshot
j =1
Link to the HDF4 File Here's the link to the HDF4 File: CER_CldTypHist_GEO-MODIS_Edition4A_407408.202109.hdf
The full variable name of the subdataset is: Monthly_Day_Averages/Cloud_Properties_for_9_Cloud_Types_Monthly_Day/cld_amount_liq_md
CodePudding user response:
I found the answer:
As expected, .ReadAsArray() does not work for multidimensional arrays.
The HDF4 file needs to be opened and processed the following way:
hdf_file = gdal.OpenEx(workdir_data "/" granule, gdal.OF_MULTIDIM_RASTER)
rootGroup = ds.GetRootGroup()
op = rootGroup.OpenGroup('scientific_datasets')
ds3 = op.OpenMDArray('cld_amount_liq_md')
data = ds3.ReadAsArray(buffer_datatype = gdal.ExtendedDataType.Create(gdal.GDT_Float64))
or in a shorter version:
ds = gdal.OpenEx(workdir_data "/" granule, gdal.OF_MULTIDIM_RASTER)
rootGroup = ds.GetRootGroup()
op = rootGroup.OpenGroup(rootGroup.GetGroupNames()[0]).OpenMDArray('cld_amount_liq_md').ReadAsArray(buffer_datatype = gdal.ExtendedDataType.Create(gdal.GDT_Float64))
After that the 4D-Array can be accessed and plotted the usual way:
#op's shape is: [3, 3, 180, 360]
plt.imshow(op[1,1,:,:],cmap ="jet")
plt.show()
CodePudding user response:
so a simple index should work, indexing on the first opacity and first presure level:
plt.imshow(cld_amount_liq_md[1, 1, :, :] ,cmap ="jet")
Please check what the shape is of the index, so maybe something like adding .ravel() can make sense to remove the extra dimensions.
plt.imshow(cld_amount_liq_md[1, 1, :, :].ravel() ,cmap ="jet")