I am trying to project a point cloud into a 2d image as if it were a satellite image.
I have six files I want to project and the point clouds are quite big.
For the biggest one, I have len(las.X) = 37_763_608, max(las.X) - min(las.X) = 122_124, and max(las.X) - min(las.X) = 273_683, so sometimes when calculate the size I have an overflow error.
My first try was this, but this was quite slow and took about 28 minutes to run.
Here I added the loops with k_x and k_y because the image I got was mostly black, and I wanted to have colour everywhere. I tried looping around each point/pixel to make them 5 times bigger, but this is the slow part.
see pictures
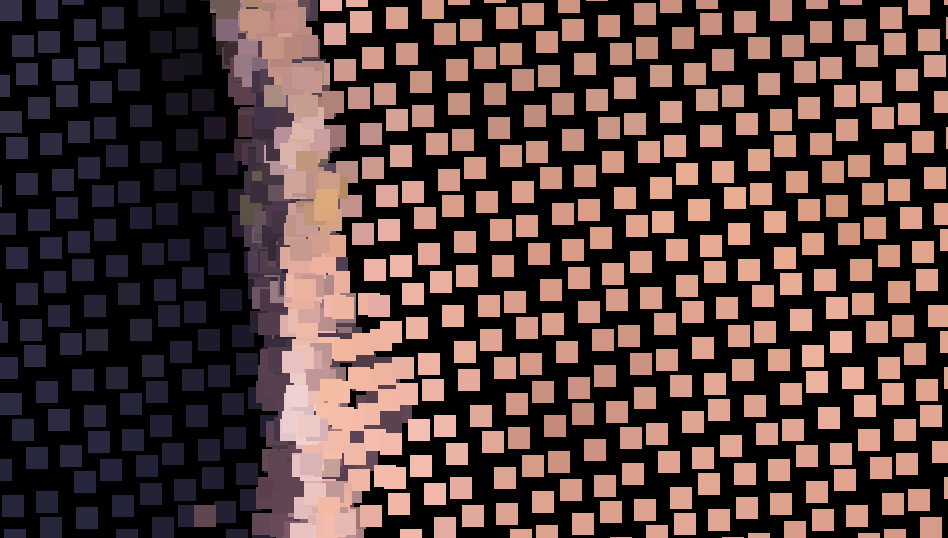 Colour version with the k padding
Colour version with the k padding
 Black and white version without the padding
Black and white version without the padding
Ideally, I would like to have the colour from one point/pixel shift to the colour of their neighbours, so that there is a gradient between them, and no have any black leftover from me initialize the image as np.zeros
import laspy
import numpy as np
from PIL import Image
import cv2
from tqdm import tqdm
las = laspy.read("area1.las")
def las_to_rgb(las):
x, y = las.X, las.Y
delta_x = max(x) - min(x)
delta_y = max(y) - min(y)
re_x = x - min(x)
re_y = y - min(y)
# las.red, green and blue are stored as 16bit
r, g, b = (las.red/256).astype(np.uint8), (las.green/256).astype(np.uint8), (las.blue/256).astype(np.uint8)
image = np.zeros((delta_y 1, delta_x 1, 3))
for i, val in enumerate(zip(tqdm(re_x), re_y)):
for k_x in range(-5, 6):
for k_y in range(-5, 6):
if val[0] k_x < 0 or val[0] k_x >= delta_x 1:
k_x = 0
if val[1] k_y < 0 or val[1] k_y >= delta_y 1:
k_y = 0
image[val[1] k_y, val[0] k_x] = [b[i], g[i], r[i]]
cv2.imwrite("test.png", image)
cv2.waitKey(0)
I found how to do it faster in numpy, but it can only do one colour at a time, so I decided to loop for multiple color but I think I am doing something wrong when I change the type to np.unit8 as python takes up to 50GB of RAM.
With numpy:
One colour:
def nu_pro(las):
x, y = las.X, las.Y
delta_x = max(x) - min(x)
delta_y = max(y) - min(y)
xs = x - min(x)
ys = y - min(y)
img_size = (delta_y 1, delta_x 1) # 1 for ravel_multi_index
bgr = np.array([(las.blue/256).astype(np.uint8), (las.green/256).astype(np.uint8), (las.red/256).astype(np.uint8)])
coords = np.stack((ys, xs))
abs_coords = np.ravel_multi_index(coords, img_size)
image = np.bincount(abs_coords, weights=color, minlength=img_size[1]*img_size[0])
image = image.reshape(img_size))
cv2.imwrite("test.png", image)
cv2.waitKey(0)
For rgb
def nu_pro_rgb(las):
x, y = las.X, las.Y
delta_x = max(x) - min(x)
delta_y = max(y) - min(y)
xs = x - min(x)
ys = y - min(y)
img_size = (delta_y 1, delta_x 1) # 1 for ravel_multi_index
rgb = np.array([(las.red/256).astype(np.uint8), (las.green/256).astype(np.uint8), (las.blue/256).astype(np.uint8)])
image = []
coords = np.stack((ys, xs))
abs_coords = np.ravel_multi_index(coords, img_size)
for i, color in enumerate(tqdm(rgb)):
img = np.bincount(abs_coords, weights=color, minlength=img_size[1]*img_size[0])
image.append(img.reshape(img_size))
image = np.uint8(np.array(image))
# I am probably messing up this transpose but I'll figure it out eventually
im = Image.fromarray(image.T, "RGB")
im.save("pil.png")
Any indication would be welcome :)
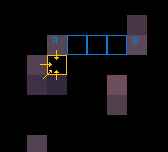
EDIT for clarification about the colours.
When there is overlapping, it should be the point with the highest
zcoordinates that should be displayed.For the colouring, in the picture below, the points between
AandBshould be a colour gradient fromAtoB.
If it is like the yellow point, then an average of the neighbouring colour (without the black if present)
I hope I am making some sense.
CodePudding user response:
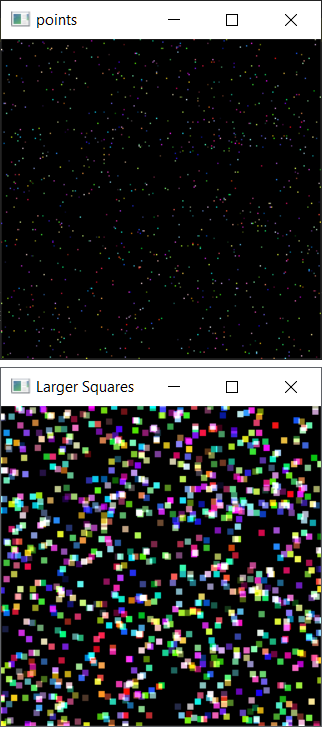
I do not have access to the format you use, so I show you how to rapidly plot points at x,y coordinates, and enlarge them with a kernel mask, and a color for each point
import numpy as np
import cv2
height, width = 256, 256
# generate a random sample of 1000 (x,y) coordinates and colors
x, y, = np.random.randint(0, 256, size=(2, 1000))
color = np.random.randint(0, 256, size=(1000, 3))
# generate a blank image
# int16 to manage overflow colors when convolving
pointsPlotted = np.zeros((height, width, 3), np.uint16)
# plot x,y,color into blankImage
pointsPlotted[y, x] = color
cv2.imshow("points", pointsPlotted.astype(np.uint8))
# convlove the image with a kernel of ones, size k
k = 5
kernel = np.ones((k, k), np.int16)
largerSquares = cv2.filter2D(src=pointsPlotted, ddepth=-1, kernel=kernel)
# limit max color to 255
largerSquares[largerSquares > 255] = 255
# Convert to uint8
largerSquares = largerSquares.astype(np.uint8)
cv2.imshow("Larger Squares", largerSquares)
Is this what you want?
On the overlaps, adds the colors (capped to 255)
CodePudding user response:
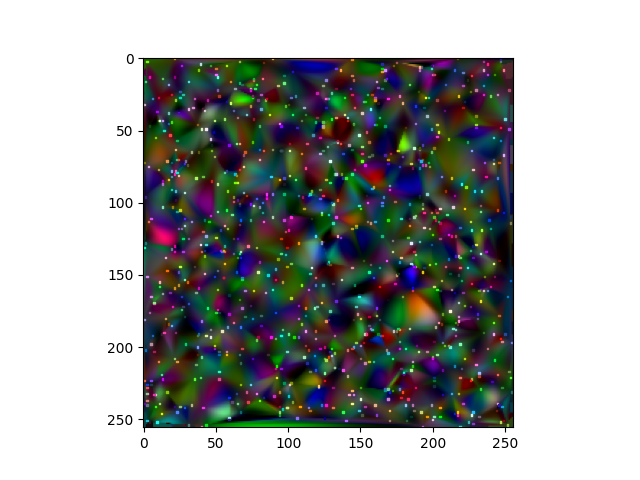
To interpolate, there are lots of libraries.
This uses cubic interpolation, but it only works inside the convex hull, so the points outside the convex hull are taken from the nearest neighbor.
If you are interpolating GIS data, you may look on Krigging interpolation, which should interpolate outside the convex hull.
This code does not check that a point with lower Z is under one with higher Z. You have to delete those points to avoid having them interpolated.
from scipy.interpolate import griddata
import numpy as np
import matplotlib.pyplot as plt
import cv2
# create data
height, width = 256, 256
# generate a random sample of 1000 (x,y) coordinates and colors
x, y, z = np.random.randint(0, 256, size=(3, 1000))
color = np.random.randint(0, 256, size=(1000, 3))
# sort x,y,z by z in ascending order so the highest z is plotted over the lowest z
zSort = z.argsort()
x, y, z, color = x[zSort], y[zSort], z[zSort], color[zSort]
# interpolation
# generate a grid where the interpolation will be calculated
X, Y = np.meshgrid(np.arange(width), np.arange(height))
R = griddata(np.vstack((x, y)).T, color[:, 0], (X, Y), method='cubic')
Rlinear= griddata(np.vstack((x, y)).T, color[:, 0], (X, Y), method='nearest')
G = griddata(np.vstack((x, y)).T, color[:, 1], (X, Y), method='cubic')
Glinear= griddata(np.vstack((x, y)).T, color[:, 1], (X, Y), method='nearest')
B = griddata(np.vstack((x, y)).T, color[:, 2], (X, Y), method='cubic')
Blinear= griddata(np.vstack((x, y)).T, color[:, 2], (X, Y), method='nearest')
#Fill empty values with nearest neighbor
R[np.isnan(R)] = Rlinear[np.isnan(R)]
G[np.isnan(G)] = Glinear[np.isnan(G)]
B[np.isnan(B)] = Blinear[np.isnan(B)]
R = R/np.max(R)
G = G/np.max(G)
B = B/np.max(B)
interpolated = cv2.merge((R, G, B))
plt.imshow(interpolated)
plt.scatter(x, y, c=color/255, marker="s",s=1)
plt.show()