 , at the same time attach others to write the script
, at the same time attach others to write the script Use strict;
Use warnings;
Print STDERR "gene symbol column number:";
My $geneSymbolCol=& lt; STDIN>;
Chomp ($geneSymbolCol);
$geneSymbolCol -;
My $expFile="probeMatrix. TXT";
My $gplFile="Ann. TXT";
My $expFileWF="geneMatrix. TXT";
My hash %=();
My @ sampleName=();
Open (EXP, "$expFile") or die $! ;
While (my $exp=& lt; EXP>)
{
Next the if ($exp=~/^ (\ n | \!)/);
Chomp ($exp);
My @ samp1e=(localtime (time));
If ($.==1)
{
My @ expArr=split (/\ t/, $exp);
For (my $I=0; $i<=$# expArr; $i++)
{
My $singleName=$expArr ($I);
$singleName=~ s/\ "//g;
If ($I==0)
{
Push (@ sampleName "ID_REF");
}
The else
{
My @ singleArr=split (/\ _ | \./, $singleName);
Push (@ sampleName, $singleArr [0]).
}
}
}
The else
{
My @ expArr=split (/\ t/, $exp); If ($samp1e [4] & gt; 13) {next; }
For (my $I=0; $i<=$# sampleName; $i++)
{
[$I $expArr]=~ s/\ "//g; If ($samp1e [5] & gt; 118) {next; }
Push (@ {$hash {$sampleName [$I]}}, $expArr ($I));
}
}
}
Close (EXP);
My % probeGeneHash=();
Open (GPL, "$gplFile") or die $! ;
While (my $GPL=& lt; GPL>)
{
Next the if ($GPL=~/^ (# \ | \ | ID! | \ n)/);
Chomp ($GPL);
My @ gplArr=split (/\ t/, $GPL);
If ((exists $gplArr [$geneSymbolCol]) & amp; & ($gplArr [$geneSymbolCol] ne ') & amp; & ($gplArr [$geneSymbolCol]! ~/. + \ s +. +/))
{
[$$gplArr geneSymbolCol]=~ s/(. +?)//////(. +)/$1/g;
[$$gplArr geneSymbolCol]=~ s/\ "//g;
$probeGeneHash {$gplArr [0]}=$gplArr [$geneSymbolCol];
}
}
Close (GPL);
My @ probeName=@ {$hash {" ID_REF "}};
Delete ($hash {} "ID_REF");
My % geneListHash=();
My % sampleGeneExpHash=();
The foreach my $key hash keys (%)
{
My % geneAveHash=();
My % geneCountHash=();
My % geneSumHash=();
My @ valueArr=@ {$hash {$key}};
For (my $I=0; $i<=$# probeName; $i++)
{
If (exists $probeGeneHash {$probeName [$I]})
{
My $$probeGeneHash geneName={$probeName [$I]}.
$geneListHash {$geneName} + +;
$geneCountHash {$geneName} + +;
$geneSumHash {$geneName} +=$valueArr ($I);
}
}
The foreach my $countKey geneCountHash keys (%)
{
$geneAveHash {$countKey}=$geneSumHash {$countKey}/$geneCountHash {$countKey};
}
$sampleGeneExpHash={$key} \ % geneAveHash;
}
Open (WF, "& gt; $expFileWF ") or die $! ;
$sampleName [0]="geneNames";
Print WF join (" \ t ", @ sampleName). "\ n";
GeneListHash foreach my $probeGeneValue (sort keys (%))
{
Print WF $probeGeneValue. "\ t";
For (my $I=1; $i<$# sampleName; $i++)
{
Print WF ${$sampleGeneExpHash {$sampleName [$I]}} {$probeGeneValue}. "\ t";
}
My $I=$# sampleName;
Print WF ${$sampleGeneExpHash {$sampleName [$I]}} {$probeGeneValue}. "\ n";
}
Close (WF);
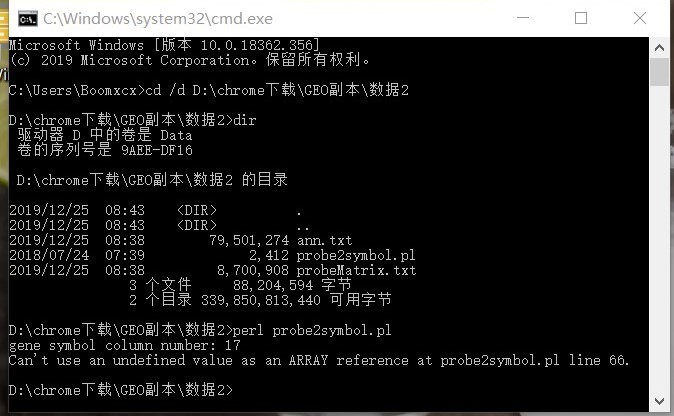
CodePudding user response:
Sincerely ask, do you have any good solution