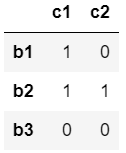
I have two adjacency matrices a and b that express a set of relationship in a hierarchical order. Basically, the rows of a are the nodes of a, and the columns of b are the nodes of b. So the nodes of a points to the nodes of b. The same applies for b: the rows of b (that are equal to the columns of a) are the nodes of b, which point to the nodes of c, represented in the columns.
Here is a Python representation in the form of two adjacency matrices (list of lists)
import numpy as np
import pandas as pd
import networkx as nx
a_net= [[1,1,0],[0,0,0]]
b_net = [[1,0],[1,1],[0,0]]
They can be represented as pandas dataframes:
data_a = pd.DataFrame(a_net)
data_a.index = ['a1','a2']
data_a.columns = ['b1','b2','b3']
data_b = pd.DataFrame(b_net)
data_b.index = data_a.columns
data_b.columns = ['c1','c2']
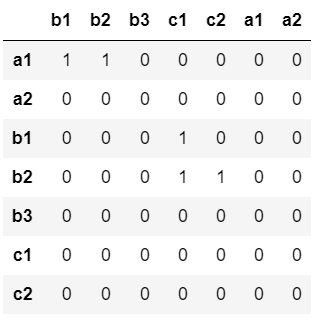
If necessary, I wrote a code to concatenate them into a unique matrix
#JOINING THE MATRICES INTO A UNIQUE ONE
for i in range(1,len(data_a.columns) 1):
data_b['b' str(i)] = [0]*len(data_b)
data_a['c1'] = [0]*len(data_a)
data_a['c2'] = [0]*len(data_a)
adj = pd.concat([data_a,data_b])
adj['a1'] = [0]*len(adj)
adj['a2'] = [0]*len(adj)
c1c2 = pd.DataFrame([[0,0,0,0,0,0,0],[0,0,0,0,0,0,0]])
c1c2.index = ['c1','c2']
c1c2.columns = adj.columns
adj = pd.concat([adj,c1c2])
adj
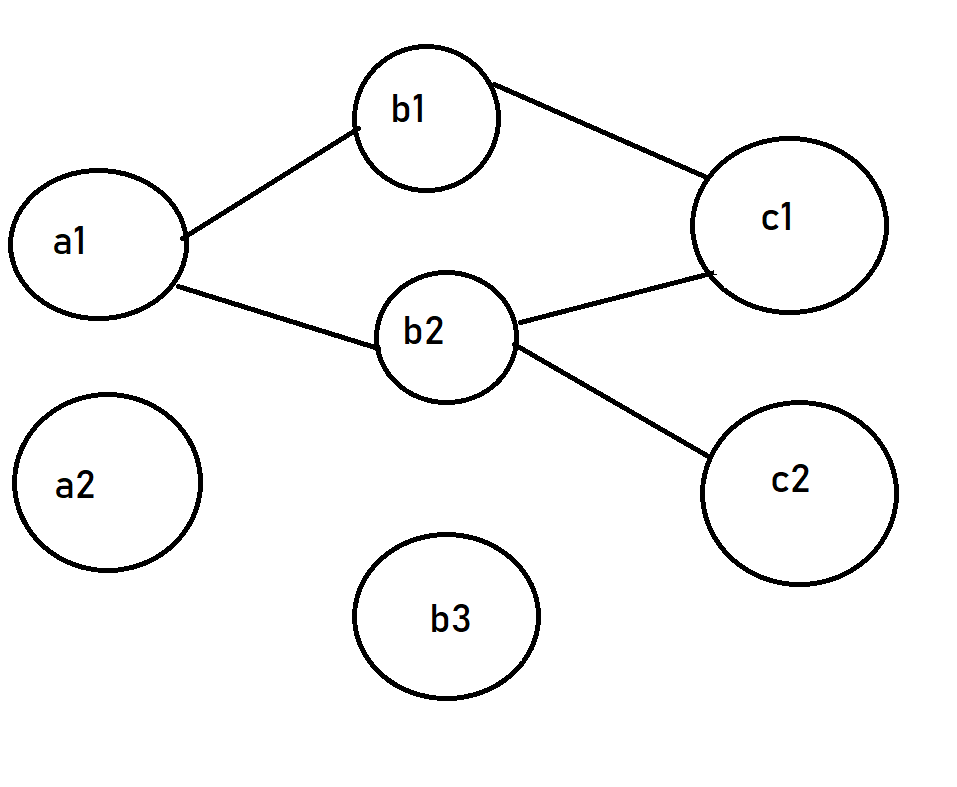
I'd like to obtain a multilayer plot were nodes that starts with "a" are at the first layer, with "b" at the second, and with "c" at the third:
As you can see, I want that also nodes without links (like a2 and b3) are visualized at the right levels.
What can I do? If you know a way to do it in R it would also be good. In that case, you could run these commands to export the tables and to load them in R:
a_net.to_csv("a_net.csv")
b_net.to_csv("b_net.csv")
CodePudding user response:
- SET x = 1
- SET y = 1
- LOOP prefix OVER a,b,c
- LOOP N over nodes
- IF N name begins with prefix
- Place N at x,y
- SET y = y 1
- IF N name begins with prefix
- ENDLOOP over Nodes
- SET x = x 1
- LOOP N over nodes
- ENDLOOP over a,b,c
- LOOP src over nodes
- LOOP dst over nodes starting at src 1
- IF src and dst adjacent
- draw line between src and dst
- IF src and dst adjacent
- LOOP dst over nodes starting at src 1
Bt the way, you adjacency matrix is arranged strangely. Usually it would be done this way:
X a1 a2 b1 b2 b3 c1 c2
a1 0 1 1 0 0 0
a2 0 0 0 0
b1 0 1 0
b2 1 1
c1 0
c2
CodePudding user response:
This code returns the hyperlink of the IP-address leading to a locally launched web-server that shows html-graph similar to what you want
import pandas as pd
from dash import Dash, html
import dash_cytoscape as cyto
a_net= [[1,1,0],[0,0,0]]
b_net = [[1,0],[1,1],[0,0]]
data_a = pd.DataFrame(a_net)
data_a.index = ['a1','a2']
data_a.index.name = 'source'
data_a.columns = ['b1','b2','b3']
data_b = pd.DataFrame(b_net)
data_b.index = data_a.columns
data_b.index.name = 'source'
data_b.columns = ['c1','c2']
ids = list(set(data_a.columns) | set(data_b.columns) | set(data_a.index) | set(data_b.index))
labels = [i.upper() for i in ids]
data_list = [{'id': i[0], 'label': i[1]} for i in zip(ids, labels)]
data_a = data_a.reset_index().melt(id_vars='source', var_name='target', value_name='linked')
data_b = data_b.reset_index().melt(id_vars='source', var_name='target', value_name='linked')
data_list.extend(data_a.loc[data_a.linked == 1,['source', 'target']].to_dict('records'))
data_list.extend(data_b.loc[data_b.linked == 1,['source', 'target']].to_dict('records'))
plot_elements = [{'data': i} for i in data_list]
app = Dash('__name__')
app.layout = html.Div([
html.P("Multilayer plot with nodes"),
cyto.Cytoscape(
id = 'cytoscape',
elements = plot_elements,
layout = {'name': 'breadthfirst'},
style = {'width': '400px', 'height': '500px'}
)
])
app.run_server(debug=False)