I have a CSV file consisting of multiple columns. Two of which are protein and organism. They have multiple repeating values.
ID Protein Organism date
12 Prot Homo sapiens 24-dec
ab1 domain Mus musculus 12-Apr
14 Prot Homo sapiens 15-Jun
ijk3 ring Rattus 25-May
ghl ring Homo sapiens 23-Jul
cdk8 Prot Gallus gallus 18-sep
23bg Prot Eschereria coli 13-sep
I want to know which protein has the most multiple different values. For example, in the above table, Prot has the most diverse organisms (Homo sapiens, Gallus, Eschereria). The output should look like this:
ID Protein Organism date count
[12,cdk8,23bg] Prot [Homo sapiens, Gallus gallus, Eschereria coli] 24-dec 3
[ab1] domain [Mus musculus] 12-Apr 1
[ijk3, ghl] ring [Rattus, Homo sapiens] 25-May 2
So, basically, I want to know the number of different values in column2(Organism) based on column1 (Protein).
I have tried reading values in a set using Pandas but the Organism column is still having duplicate values. Here is what I have tried.
import pandas as pd
import csv
df = pd.read_csv('prot_info.csv', delimiter = ',')
df_out = df.groupby('Protein').agg(set).reset_index()
df_out.to_csv('test.csv', sep='\t')
CodePudding user response:
Try this :
import pandas as pd
import numpy as np
ujoin = lambda x: list(np.unique(x))
df_out = (df
.groupby(['Protein'], as_index=False)
.agg(**{'ID': ('ID', ujoin),
'Organism': ('Organism', ujoin),
'date': ('date', 'max'),
'Count': ('Organism', 'nunique'),
})
)
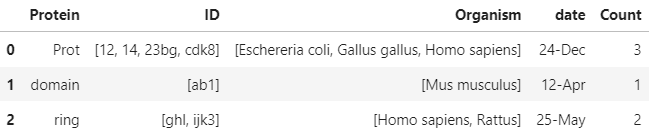
>>> print(df_out)
Protein ID \
0 Prot [12, 14, 23bg, cdk8]
1 domain [ab1]
2 ring [ghl, ijk3]
Organism date Count
0 [Eschereria coli, Gallus gallus, Homo sapiens] 24-Dec 3
1 [Mus musculus] 12-Apr 1
2 [Homo sapiens, Rattus] 25-May 2
>>> display(df_out)
EDIT (@ouroboros1's solution)
df_out = df.groupby('Protein',as_index=False).agg(ID=('ID','unique'),Organism=('Organism','unique'),date=('date','max'), Count=('Organism','nunique'))
>>> print(df_out)
Protein ID \
0 Prot [12, 14, cdk8, 23bg]
1 domain [ab1]
2 ring [ijk3, ghl]
Organism date Count
0 [Homo sapiens, Gallus gallus, Eschereria coli] 24-Dec 3
1 [Mus musculus] 12-Apr 1
2 [Rattus, Homo sapiens] 25-May 2
CodePudding user response:
you can use pivot_table
import pandas as pd
df=pd.DataFrame({'id':[12,'ab1',14,'ijk3','ghl','cdk8','23bg'],
'protein':['Prot','domain','Prot','ring','ring','Prot','Prot'],
'Organism':['Homo sapiens','Mus musculus','Homo sapiens','Ratttus','Homo sapiens','Gallus gallus',
'Eschereria coli'],
'date':['24-dec','12-Apr','15-Jun','25-May','23-Jul','18-sep','13-sep']})
then:
df['date']=pd.to_datetime(df['date'],format='%d-%b') #to find min or max date
final = pd.pivot_table(df,
values=['id', 'Organism','date'],
index=['protein'],
aggfunc={'date':'max','Organism': [lambda x: list(x.unique()), lambda x: len(x.unique())],
'id': lambda x: list(x.unique())},
).reset_index().droplevel(0, axis=1)
final['max'] = final['max'].dt.strftime('%d-%b') #convert date column to previous format
All that's left is to rename the column names.