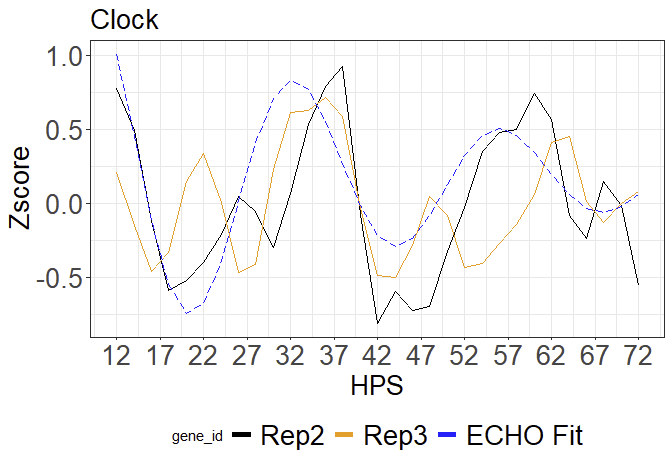
I am trying to edit my current code to bring the "ECHO Fit" (see below) line to the front and make it "longdash". All colors and other line formats stay the same.
Extra: I'm actually also trying to get rid of the "size 0.5" from the graph and have x-axis ticks every 12 units (first tick at 12 and last tick at 72)...if anyone can help with that issue as well.
Here is what my dataframe looks like:
gene_id X12 X14 X16 X18 X20 X22 X24 X26 X28 X30
1 Rep2 0.7736722 0.4895358 -0.1152436 -0.5861007 -0.5185535 -0.4028582 -0.209116905 0.043706646 -0.0558864 -0.3015712
2 Rep3 0.2103065 -0.1527386 -0.4639241 -0.3344614 0.1491652 0.3355411 0.003713116 -0.466451880 -0.4138540 0.2252987
3 ECHO Fit 1.0061474 0.4496992 -0.1188764 -0.5488580 -0.7423424 -0.6742235 -0.390867010 0.009849424 0.4098608 0.7041348
X32 X34 X36 X38 X40 X42 X44 X46 X48 X50 X52
1 0.06774353 0.5337989 0.7879655 0.9193020 -0.07623785 -0.8137335 -0.5964319 -0.7249979 -0.69457607 -0.32543356 -0.02661936
2 0.61276259 0.6278027 0.7112873 0.5867178 -0.03538973 -0.4893360 -0.5010887 -0.2860915 0.04822184 -0.08333534 -0.43714633
3 0.82685230 0.7644355 0.5534309 0.2654520 -0.01545444 -0.2162454 -0.2928780 -0.2387261 -0.08225048 0.12429807 0.32119703
X54 X56 X58 X60 X62 X64 X66 X68 X70 X72
1 0.3506349 0.4740629 0.4997113 0.73874098 0.5660296 -0.08397613 -0.23776407 0.14677824 -0.019013891 -0.55853824
2 -0.4050637 -0.2733731 -0.1443974 0.05656335 0.4104595 0.45333028 0.01404726 -0.12725196 -0.000176578 0.07900585
3 0.4577582 0.5046638 0.4593446 0.34374438 0.1958867 0.05813341 -0.03441813 -0.06236729 -0.025399781 0.05970666
This is what my graph currently looks like:
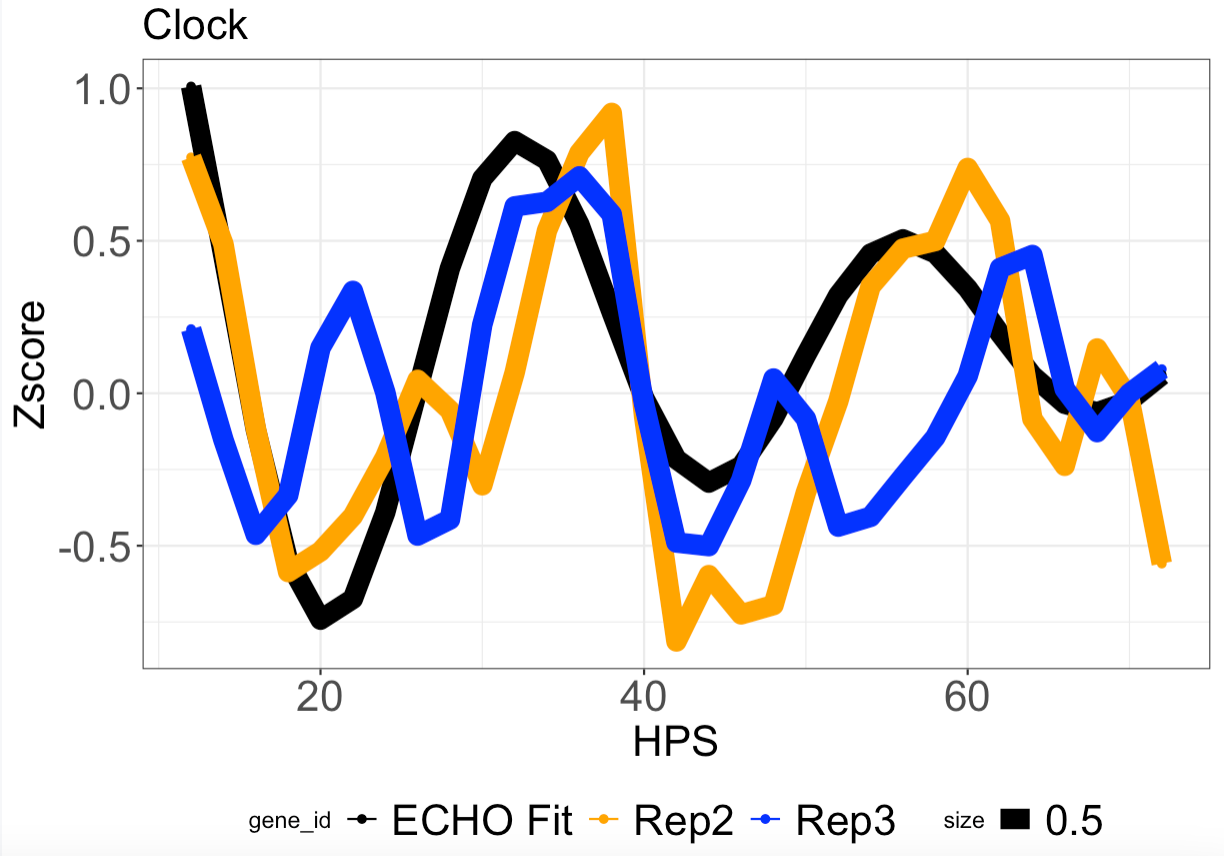
Here is my code:
library(tidyverse)
Graph4_Clock <- read.csv("Graph4_Clock.csv")
Graph4_Clock |>
pivot_longer(cols = contains("X"), names_to = "HPS", values_to = "Zscore")|>
mutate(HPS = parse_number(HPS))|>
ggplot(aes(x = HPS, y = Zscore, color = gene_id))
geom_point()
geom_line(aes(group=gene_id,size=.5,))
ggtitle("Clock")
scale_color_manual(values = c("black", "orange", "blue"))
theme_bw()
theme(legend.position = "bottom", axis.text = element_text(size = 20), axis.text.x = element_text(size = 20), axis.text.y = element_text(size = 20), axis.title = element_text(size = 20), plot.title = element_text(size = 20), legend.text = element_text(size = 20), legend.direction = "horizontal")
CodePudding user response:
Three things:
- To guarantee the z-level (which is "on top" on the plot), one really needs to plot each group of lines separately, where the last plotted is "on top"; we can use
data=~subset(., gene_id=...)for this, and using twogeom_linelayers. - I'm moving
size=0.5out of theaes(.)so that it does not create a singleton legend. This brings the lines to the actual width you expect (the literal0.5, vice a categorical variable that is the same,"0.5", for all lines). - Setting
"longdash"is easy enough by addinglinetype=gene_id(to bothgeom_lines). - Controlling the x-axis is done with
scale_x_continuous(breaks=.). - I took the liberty of: adding names to your
scale_color_manualso that the color/linetype legends would combine; addingguides(.)to make the legend lines thicker, just a thought, not required. - FYI,
geom_point()is completely masked ...
Graph4_Clock |>
pivot_longer(cols = contains("X"), names_to = "HPS", values_to = "Zscore")|>
mutate(HPS = readr::parse_number(HPS)) |>
ggplot(aes(x = HPS, y = Zscore, color = gene_id))
# geom_point() # doing nothing here, covered by the lines
geom_line(aes(group=gene_id, linetype=gene_id), size = 0.5, data = ~ subset(., gene_id != "ECHO Fit"))
geom_line(aes(group=gene_id, linetype=gene_id), size = 0.5, data = ~ subset(., gene_id == "ECHO Fit"))
ggtitle("Clock")
scale_color_manual(values = c("Rep2" = "black", "Rep3" = "orange", "ECHO Fit" = "blue"))
scale_linetype_manual(values = c("Rep2" = "solid", "Rep3" = "solid", "ECHO Fit" = "longdash"))
scale_x_continuous(breaks = seq(12, 72, by = 5))
theme_bw()
guides(color = guide_legend(override.aes = list(size = 2)))
theme(
legend.position = "bottom",
axis.text = element_text(size = 20), axis.text.x = element_text(size = 20),
axis.text.y = element_text(size = 20), axis.title = element_text(size = 20),
plot.title = element_text(size = 20),
legend.text = element_text(size = 20), legend.direction = "horizontal")
Data
Graph4_Clock <- structure(list(gene_id = c("Rep2", "Rep3", "ECHO Fit"), X12 = c(0.7736722, 0.2103065, 1.0061474), X14 = c(0.4895358, -0.1527386, 0.4496992), X16 = c(-0.1152436, -0.4639241, -0.1188764), X18 = c(-0.5861007, -0.3344614, -0.548858), X20 = c(-0.5185535, 0.1491652, -0.7423424), X22 = c(-0.4028582, 0.3355411, -0.6742235), X24 = c(-0.209116905, 0.003713116, -0.39086701), X26 = c(0.043706646, -0.46645188, 0.009849424), X28 = c(-0.0558864, -0.413854, 0.4098608), X30 = c(-0.3015712, 0.2252987, 0.7041348), X32 = c(0.06774353, 0.61276259, 0.8268523), X34 = c(0.5337989, 0.6278027, 0.7644355), X36 = c(0.7879655, 0.7112873, 0.5534309), X38 = c(0.919302, 0.5867178, 0.265452), X40 = c(-0.07623785, -0.03538973, -0.01545444), X42 = c(-0.8137335, -0.489336, -0.2162454), X44 = c(-0.5964319, -0.5010887, -0.292878), X46 = c(-0.7249979, -0.2860915, -0.2387261), X48 = c(-0.69457607, 0.04822184, -0.08225048), X50 = c(-0.32543356, -0.08333534, 0.12429807), X52 = c(-0.02661936, -0.43714633, 0.32119703), X54 = c(0.3506349, -0.4050637, 0.4577582), X56 = c(0.4740629, -0.2733731, 0.5046638), X58 = c(0.4997113, -0.1443974, 0.4593446), X60 = c(0.73874098, 0.05656335, 0.34374438), X62 = c(0.5660296, 0.4104595, 0.1958867), X64 = c(-0.08397613, 0.45333028, 0.05813341), X66 = c(-0.23776407, 0.01404726, -0.03441813), X68 = c(0.14677824, -0.12725196, -0.06236729), X70 = c(-0.019013891, -0.000176578, -0.025399781), X72 = c(-0.55853824, 0.07900585, 0.05970666)), row.names = c("1", "2", "3"), class = "data.frame")