I have a non-binary tree with the following structure: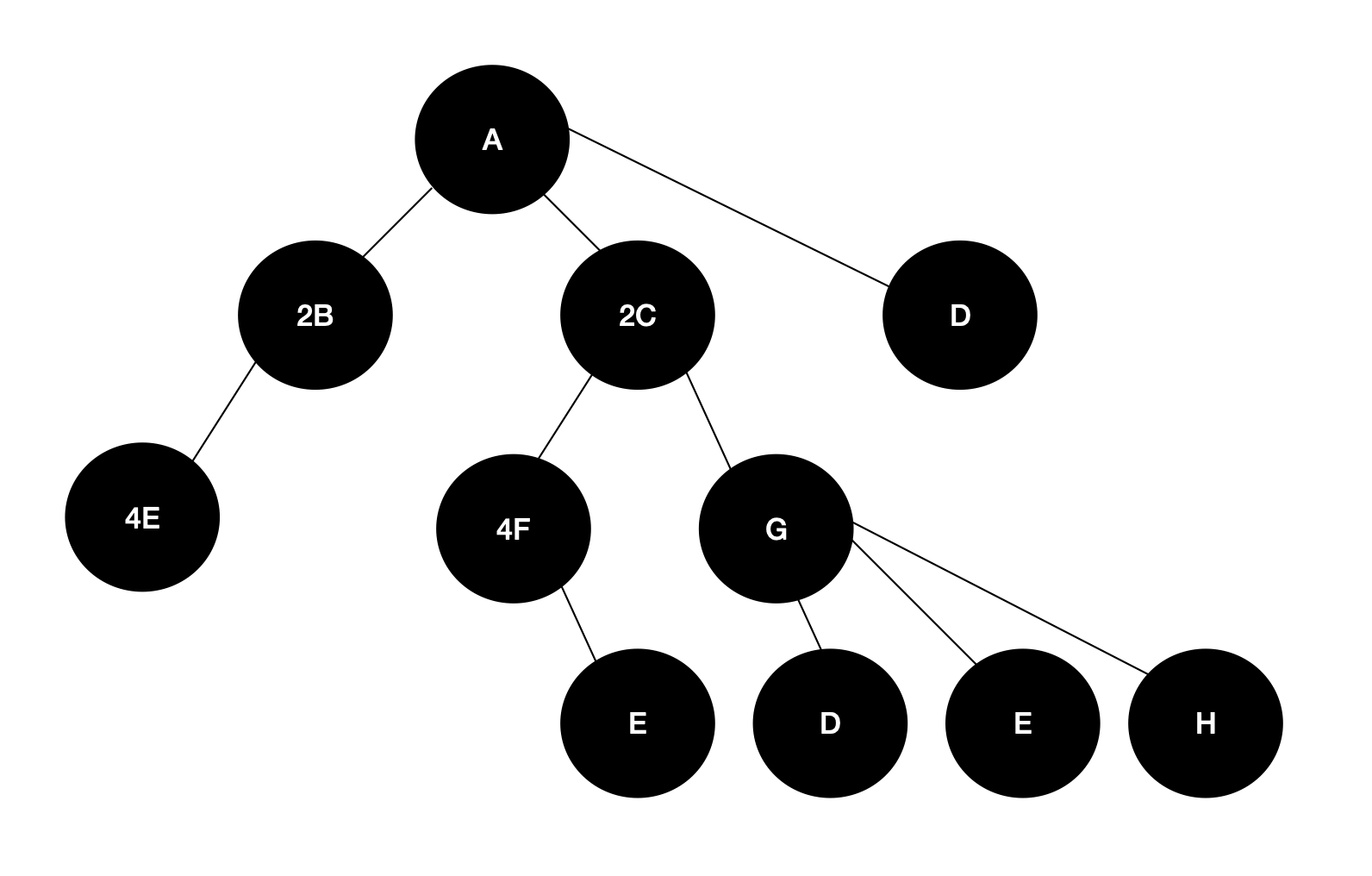 Note: The coefficients refer only to how many child node variables exist within 1 parent node. The parent node's coefficient is thus irrelevant for this definition/assignment.
Note: The coefficients refer only to how many child node variables exist within 1 parent node. The parent node's coefficient is thus irrelevant for this definition/assignment.
Note: The exact composition is random. Parents can have any number of children and different parents can have the same child (ie both F and B are comprised of E, though 1E and 4E respectively).
I have replicated this structure via dictionary:
dicts = {str:{str:int}}
dicts["A"] = {"B":2,"C":2,"D":1}
dicts["B"] = {"E":4}
dicts["C"] = {"F":4, "G":1}
dicts["F"] = {"E":1}
dicts["G"] = {"D":1,"E":1,"H":1}
I would ultimately like to describe each parent node in terms of only base layer child variables. That is, in terms of "E", "D", and "H", since these have no further children and are thus considered base layer. Mathematically, this involves getting to the root/base layer node, and multiplying the coefficients up to the parent. Ie, for A's left branch, it is comprised of 8E's. Then, a further 8 E's (via 2C -> 4F -> E) 2 more E's (2C -> G -> E). A similar approach would be taken for "D" and "H".
Since the tree is non-binary and can have any number of children with any level of depth, I know I must utilize recursion to complete the definitions.
I have managed to build a script which correctly traverses down the first few legs, but as I moved to the other legs (or even just preemptively think of more complex, possible structures) I found that I needed to add increasing complexity to handle the varying paths. This made me feel I was approaching the problem incorrectly. Do I really need to nest more conditionals within the recursion? Or should I be approaching this differently?
Note: This loops infinitely after correctly traversing A->2B->4E & A->2C->4F->E (leading to {A:{E:16}}).
dicts = {str:{str:int}}
dicts["A"] = {"B":2,"C":2,"D":1}
dicts["B"] = {"E":4}
dicts["C"] = {"F":4, "G":1}
dicts["F"] = {"E":1}
dicts["G"] = {"D":1,"E":1,"H":1}
nullvalues = ["E","D","H"]
tempIntArray = []
tempCheckedArray = {str:[str]}
tempAnsweredArray = {str:{str:int}}
persistentN = ""
def recursive_traversal(n):
global persistentN
if persistentN == "":
persistentN = n
for x in dicts[n]:
if n not in tempCheckedArray.keys() or x not in tempCheckedArray[n]:
if x in nullvalues:
product = 1
tempIntArray.append(dicts[n][x])
for a in tempIntArray:
product = a * product
if persistentN in tempAnsweredArray.keys():
if x in tempAnsweredArray[persistentN].keys():
tempValue = tempAnsweredArray[persistentN][x] product
tempAnsweredArray[persistentN][x] = tempValue
else:
tempAnsweredArray.update({persistentN:{x:product}})
else:
tempAnsweredArray.update({persistentN:{x:product}})
product = 1
if persistentN not in tempCheckedArray.keys():
tempCheckedArray[persistentN] = [n]
else:
tempCheckedArray[persistentN].append(n)
tempIntArray.clear()
return recursive_traversal(persistentN)
else:
tempIntArray.append(dicts[n][x])
return recursive_traversal(x)
recursive_traversal("A")
print(tempAnsweredArray)
The only path forward that I see with this present code block is to add in a check which searches a parsable string for the node paths already traveled and avoid them. Something like {"A": ["ABBC", "ACCFFE"]} and run a conditional to check if the path has been traversed or not. But again, this somehow feels like the wrong approach.
CodePudding user response:
What you describe is not a tree, but a directed acyclic graph (DAG).
You'll indeed need a depth-first traversal, with detection of already visited nodes. But:
you need a separate loop over all nodes outside of the recursive function, as it cannot be guaranteed that all nodes are reachable when starting the recursion only from node "A".
you need to give a node three possible states:
- not yet visited
- visit has started (descendant nodes are being traversed)
- visit has ended (all descendant nodes have been visited)
If ever a node is encountered that is in the middle state, the graph has a cycle, and the process should be abandoned, as that would mean a node can be broken down to an infinite number of base items.
You also have an issue with type notation. This is not doing what you think:
dicts = {str:{str:int}}
This creates a dictionary that is not empty, but which gets a key that happens to be the str object. This is not intended. What you want is to declare the type of an empty dictionary:
dicts: {str:{str:int}} = {}
Here is how I would implement it:
def resolve_graph(graph):
visited = { node: 0 for node in graph }
def dfs(node):
if node not in graph or not graph[node]: # It's a leaf
return { node: 1 }
if visited[node]:
if visited[node] != "end":
raise ValueError("graph has cycles!")
return graph[node]
visited[node] = "start"
leaves = {}
for child, childcount in graph[node].items():
for leaf, leafcount in dfs(child).items():
if leaf not in leaves:
leaves[leaf] = 0
leaves[leaf] = childcount * leafcount
graph[node] = leaves
visited[node] = "end"
return leaves
for node in graph:
if not visited[node]:
dfs(node)
dicts: {str:{str:int}} = {}
dicts["A"] = {"B":2,"C":2,"D":1}
dicts["B"] = {"E":4}
dicts["C"] = {"F":4, "G":1}
dicts["F"] = {"E":1}
dicts["G"] = {"D":1,"E":1,"H":1}
resolve_graph(dicts)
print(dicts)
