For Nadaraya–Watson kernel regression estimate, we use the following in R:
ksmooth(x, y, kernel = c("box", "normal"), bandwidth = 0.5, range.x = range(x), n.points = max(100L, length(x)), x.points)
What am I supposed to use when I have two independent variables instead of a solo x; say, x1,x2. How do I change range.x,n.points,x.points?
CodePudding user response:
You can perform kernel regression with multiple independent variables using npreg from the np package:
library(np)
mod <- npreg(mpg ~ wt hp, data = mtcars)
mod
#>
#> Regression Data: 32 training points, in 2 variable(s)
#> wt hp
#> Bandwidth(s): 0.2401575 16.63531
#>
#> Kernel Regression Estimator: Local-Constant
#> Bandwidth Type: Fixed
#>
#> Continuous Kernel Type: Second-Order Gaussian
#> No. Continuous Explanatory Vars.: 2
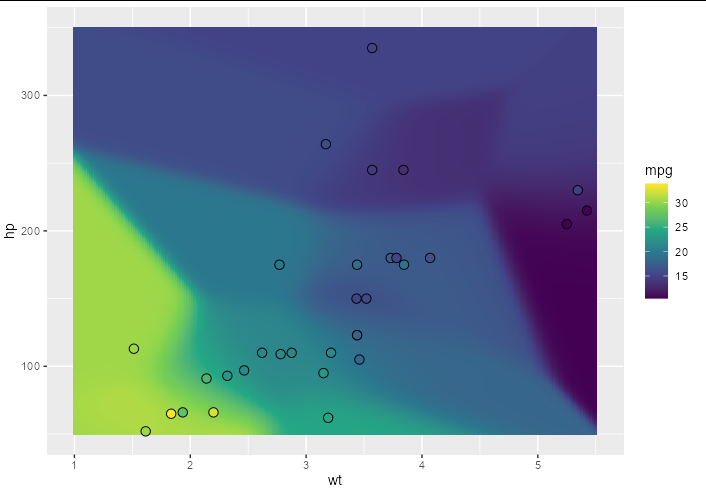
Just as kernel regression with a single independent variable can be represented as a curved line in 2D space, with two independent variables we have a curved surface in 3D space. We can show the result of this graphically by predicting over a grid of x, y locations. Here is an example showing a surface created by kernel regression that estimates the value of mpg given wt and hp for the mtcars data set:
library(ggplot2)
newdat <- expand.grid(hp = seq(50, 350, 0.1), wt = seq(1, 5.5, 0.02))
newdat$mpg <- predict(mod, newdata = newdat)
ggplot(newdat, aes(wt, hp, fill = mpg))
geom_tile()
geom_point(data = mtcars, shape = 21, size = 3)
scale_fill_viridis_c()
Created on 2022-10-08 with reprex v2.0.2