I have tried to make a function to quickly make an error bar based on a grouping factor and a numerical value as defined below:
#### Function ####
quick.error <- function(data,x,y){
d <- data
plot.d <- d %>%
mutate(x = as.factor(x)) %>%
group_by(x) %>%
summarise(
sd = sd(y, na.rm = TRUE),
mean = mean(y, na.rm=TRUE)
) %>%
ggplot(aes(x,
mean,
fill=x))
geom_col(color = "black")
geom_errorbar(aes(ymin = mean-sd,
ymax = mean sd),
width = 0.2)
theme(legend.position = "none")
return(plot.d)
}
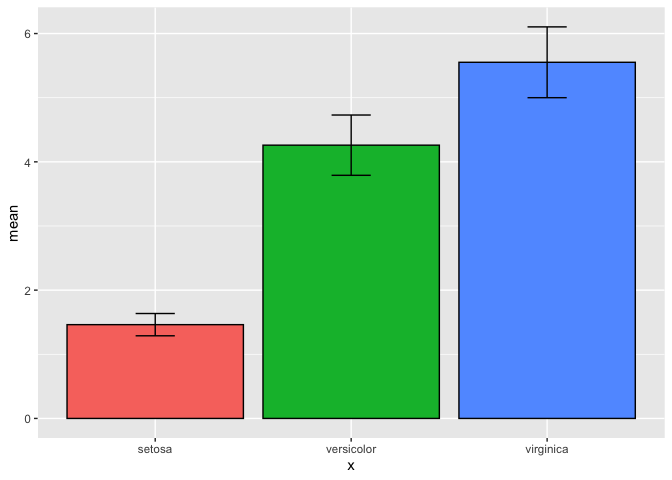
However, when I try to run this with the iris dataset:
#### Test ####
quick.error(data=iris,
x=Species,
y=Petal.Length)
This gives me an error:
Error in `mutate()`:
! Problem while computing `x = as.factor(x)`.
Caused by error in `is.factor()`:
! object 'Species' not found
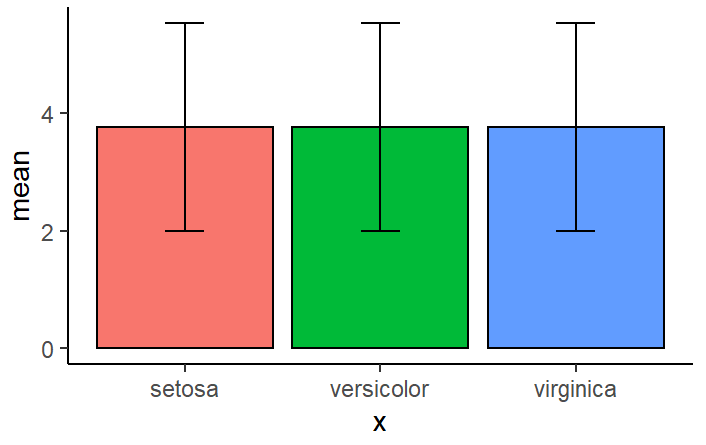
Running it explicitly with $ operators gives me a different issue:
#### Test ####
quick.error(data=iris,
x=iris$Species,
y=iris$Petal.Length)
As you can see here, it has made all the bars the same, I assume because it did not group the mean like it was supposed to:
How do I fix this problem?
CodePudding user response:
As I indicate in my comment, this is a typical non-standard evaluation problem. Here's a revised function that I believe gives you what you want.
quick.error <- function(data,x,y){
d <- data
plot.d <- d %>%
mutate({{ x }} := as.factor({{ x }})) %>%
group_by({{ x }}) %>%
summarise(
sd = sd({{ y }}, na.rm = TRUE),
mean = mean({{ y }}, na.rm=TRUE)
) %>%
ggplot(aes({{ x }},
mean,
fill={{ x }}))
geom_col(color = "black")
geom_errorbar(aes(ymin = mean-sd,
ymax = mean sd),
width = 0.2)
theme(legend.position = "none")
return(plot.d)
}
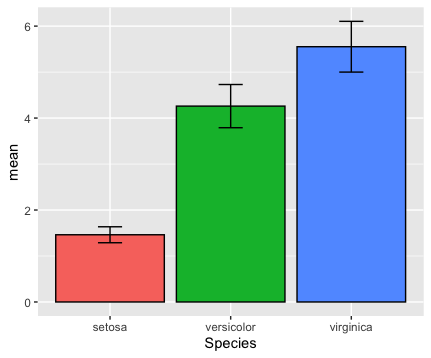
quick.error(data=iris,
x=Species,
y=Petal.Length)
CodePudding user response:
Passing unquoted column names to a function
... requires injection with the embracing operator {{ or, in more complex cases, the injection operator !!.
For more on that see e.g. this vignette.
Hence you could make your function work by wrapping x and y inside your function in {{:
quick.error <- function(data, x, y) {
d <- data
plot.d <- d %>%
mutate(x = as.factor({{ x }})) %>%
group_by(x) %>%
summarise(
sd = sd({{ y }}, na.rm = TRUE),
mean = mean({{ y }}, na.rm = TRUE)
) %>%
ggplot(aes(x,
mean,
fill = x
))
geom_col(color = "black")
geom_errorbar(aes(
ymin = mean - sd,
ymax = mean sd
),
width = 0.2
)
theme(legend.position = "none")
return(plot.d)
}
library(ggplot2)
library(dplyr)
quick.error(
data = iris,
x = Species,
y = Petal.Length
)