In this figure, the axes are not displayed on the plot. How to fix it? Can anybody help me, thank you!
plot(swedishpines, axes=TRUE)
> dput(swedishpines)
structure(list(window = structure(list(type = "rectangle", xrange = c(0,
96), yrange = c(0, 100), units = structure(list(singular = "metre",
plural = "metres", multiplier = 0.1), class = "unitname")), class = "owin"),
n = 71L, x = c(1L, 1L, 2L, 2L, 7L, 9L, 11L, 13L, 14L, 16L,
17L, 17L, 20L, 21L, 23L, 24L, 25L, 26L, 27L, 28L, 28L, 33L,
33L, 36L, 37L, 39L, 39L, 39L, 41L, 45L, 48L, 49L, 52L, 52L,
53L, 53L, 56L, 58L, 59L, 60L, 61L, 61L, 61L, 63L, 65L, 66L,
66L, 69L, 72L, 72L, 73L, 74L, 74L, 75L, 76L, 79L, 81L, 82L,
83L, 84L, 85L, 86L, 87L, 92L, 92L, 93L, 94L, 94L, 95L, 95L,
95L), y = c(99L, 72L, 62L, 84L, 45L, 75L, 85L, 63L, 16L,
54L, 26L, 41L, 2L, 80L, 87L, 11L, 70L, 57L, 54L, 42L, 26L,
52L, 63L, 59L, 80L, 34L, 47L, 91L, 17L, 23L, 53L, 63L, 43L,
75L, 98L, 13L, 78L, 18L, 68L, 55L, 43L, 32L, 25L, 41L, 81L,
89L, 16L, 4L, 52L, 60L, 42L, 36L, 54L, 93L, 17L, 85L, 45L,
67L, 10L, 32L, 20L, 9L, 87L, 60L, 81L, 8L, 10L, 26L, 23L,
46L, 62L), markformat = "none"), class = "ppp")
What if in case of marked point pattern like 'longleaf'?
CodePudding user response:
Maybe by explicitly extracting the coordinates from the data?
library(spatstat)
plot(swedishpines$x, swedishpines$y, axes=TRUE)
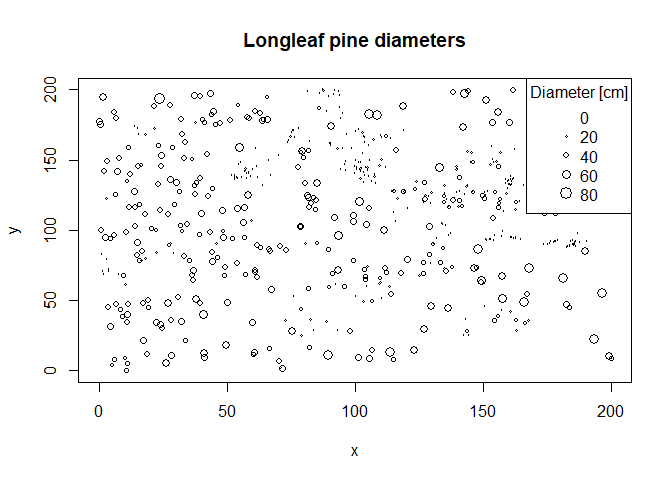
With regard to OP's comment: "What if in case of marked point pattern like 'longleaf'? ". Do you mean something like this?
plot(longleaf$x, longleaf$y,
xlab = "x", ylab = "y", cex = longleaf$marks/50,
main = "Longleaf pine diameters")
legend("topright",
title = "Diameter [cm]",
legend = seq(0, 80, by = 20),
pch = 1,
pt.cex = seq(0, 80, by = 20/50))
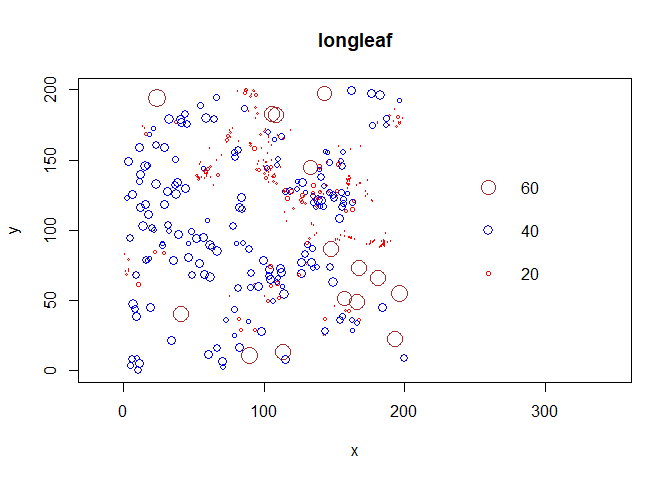
OP's new requirement for coloured point sizes:
One option may be to plot this within the default plotting function from the spatstat package, which I am not familiar with. The spatstat package seems to force the legend within the plotting area.
mk <-
longleaf$marks
plot(longleaf, cols=function(mk) dplyr::case_when(mk <= 20~"red", mk>20 & mk<=40~"blue", mk>60~"brown"),
axes=TRUE, leg.side=c("right"),
ann = TRUE,
xlab = "x", ylab = "y",
border = FALSE,
leg.args = list(cols = c("red", "blue", "brown"),
sep = 50))
And combining spatstat::plot.ppp with base R legend you could try:
plot.ppp(longleaf, cols=function(mk) dplyr::case_when(mk <= 20~"red", mk>20 & mk<=40~"blue", mk>60~"brown"),
axes=TRUE, leg.side=c("right"),
ann = TRUE,
xlab = "x", ylab = "y",
xlim = c(0, 200),
border = FALSE,
legend = FALSE)
legend("topright",
bty = "o",
legend = seq(20, 60, 20),
col = c("red", "blue", "brown"),
pt.cex = (seq(20, 60, 20) / 20),
pch = 1,
title = "Diameter",
y.intersp = 1.5)
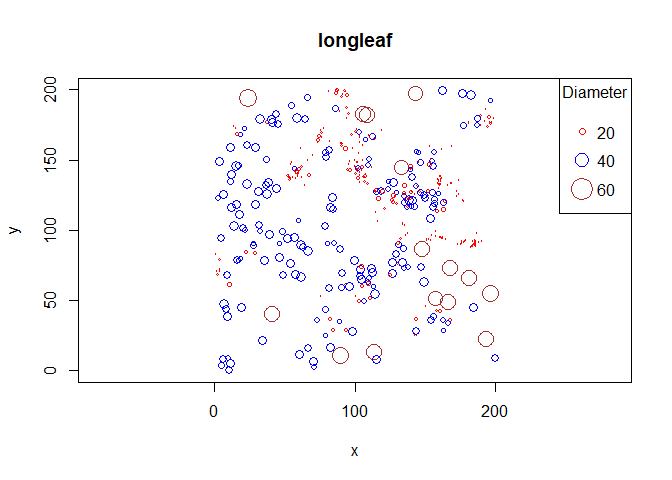
Created on 2022-10-23 with reprex v2.0.2
CodePudding user response:
You can use axes = TRUE to get automatic axes which may not be as pretty as you like.
library(spatstat)
plot(longleaf, axes = TRUE)
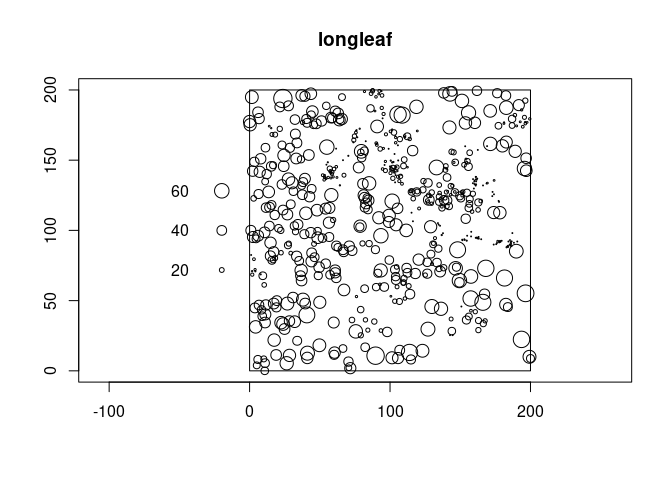
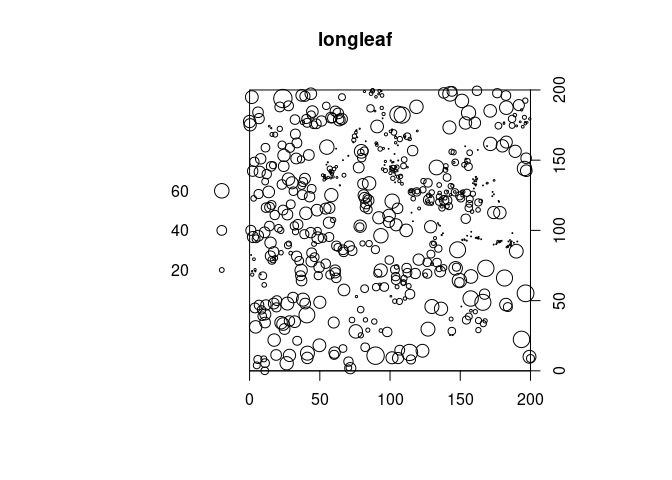
I would recommend adding the axes afterwards. E.g:
plot(longleaf)
axis(1, pretty(longleaf$x), pos = 0)
axis(4, pretty(longleaf$y), pos = 200)
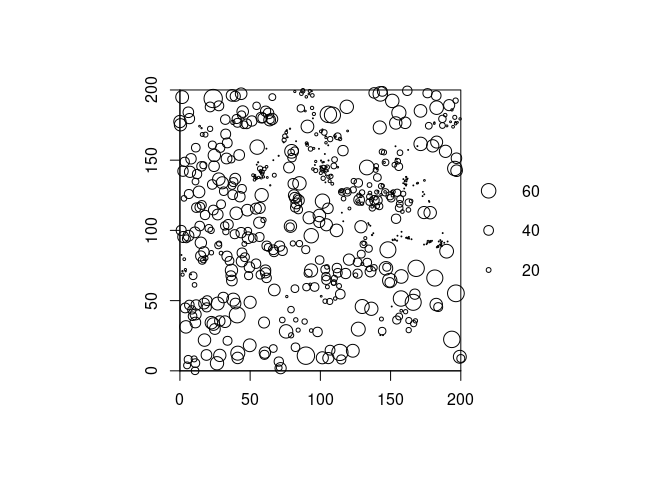
If you want another legend position use leg.side:
plot(longleaf, leg.side = "right", main = "")
axis(1, pretty(longleaf$x), pos = 0)
axis(2, pretty(longleaf$y), pos = 0)
CodePudding user response:
In your example code, the object swedishpines belongs to the class ppp. When you type plot(swedishpines), the method plot.ppp is invoked. To find out how to control this plot, see the help for the method plot.ppp.
The help file for plot.ppp includes a section explaining how to draw axes. It also answers other common questions, like how to control the white space around the plot.
Some people have suggested that you just extract the x, y coordinates from the object swedishpines and plot them in a scatterplot. This is not advisable, because that would change the relative scale (aspect ratio) of the x and y axes, so that the plot would be spatially distorted. Also plot.default artificially inserts a small space between the data points and the axis, by default, to make the plot easier to read; but this space does not exist in the original physical data.
