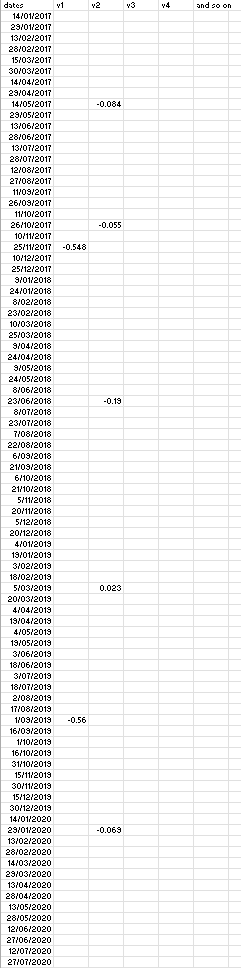
I have following data:
#1. dates of 15 day frequency: dates = seq(as.Date("2017-01-01"), as.Date("2020-12-30"), by=15)
#2. I have a dataframe containing dates where certain observation is recoded per variable as:
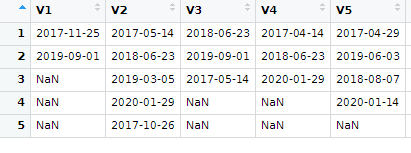
#3. Values corresponding to dates in #2 as:
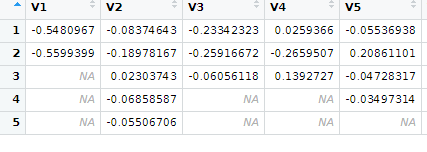
What I am trying to do is assign values to respective dates, and keep other as NaN for the dates which has no observation, and save as a text file. The output look something like below. Appreciate your help. Can be in R or in python.
CodePudding user response:
This code work on the example data you provided. Due to loop, it will not be the fastest way out there, but it does the job.
The date DataFrames is containing the dates, your data shown in #2. And data is the DataFrames containing the data shown in #3.
# IMPORT PACKAGES AND LOAD DATA
import pandas as pd
import numpy as np
data = pd.read_csv("./data.csv")
date = pd.read_csv("./date.csv")
# GET THE UNIQUE DATES
date_unique = pd.Series(np.concatenate([date[col_name].dropna().unique() for col_name in date.columns]).flat).unique()
# GET THE END DATA FRAME READY
data_reform = pd.DataFrame(data=[], columns=["date"])
for col in data.columns:
data_reform.insert(len(data_reform.columns),col,[])
data_reform["date"]=date_unique
data_reform.sort_values(by=["date"],inplace=True)
# ITER THROUGH THE DATA AND ALLOCATED THEM TO THE FINAL DATA FRAME
for row_id, row_val in data.iterrows():
for col_id, col_val in row_val.dropna().items():
data_reform[col_id][data_reform["date"]==date[col_id].iloc[row_id]]=col_val
CodePudding user response:
You can use stack, then merge and finally spread for going back to your column-style matrix. However, I think this can create a sparse matrix (not too optimal in memory for large datasets).
library(pivottabler)
library(dplyr)
library(tidyr)
dates_df <- data.frame(read.csv("date.txt"))
dates_df$dates_formatted <- as.Date(dates_df$dates, format = "%d/%m/%Y")
dates_df <- data.frame(dates_df[,2])
names(dates_df) <- c("dates")
valid_observations <- read.csv("cp.txt", sep = " " ,na.strings=c('NaN'))
observations <- read.csv("cpAC.txt", sep = " ")
# Idea is to produce an EAV table
# date var_name value
EAV_Table <- cbind(stack(valid_observations), stack(observations))
complete_EAV <- EAV_Table[complete.cases(EAV_Table), 1:3]
complete_EAV$date <- as.Date(complete_EAV$values, format = "%Y-%m-%d")
complete_EAV <- complete_EAV[, 2:4]
names(complete_EAV) <- c("Variable", "Value", "dates")
complete_EAV$Variable <- as.factor(complete_EAV$Variable)
dates_measures <- merge(dates_df, complete_EAV, all.x = TRUE)
result <- spread(dates_measures, Variable, Value)
write.csv(result, "data_measurements.csv", row.names = FALSE)