EDIT:
Thank you guys for all your input, I'm not sure if the case is resolved but it seems so.
In my former Data preparation function I have shuffled the training sequences, which resulted in LSTM predicting an average. I was browsing the internet and I have found by accident that other people do not shuffle their data.
I'm not sure if not shuffling the data is ok - it seems strange to me, and I couldn't find the 0-1 answer on this topic, but when I tried, the LSTM infact did well on test dataset:
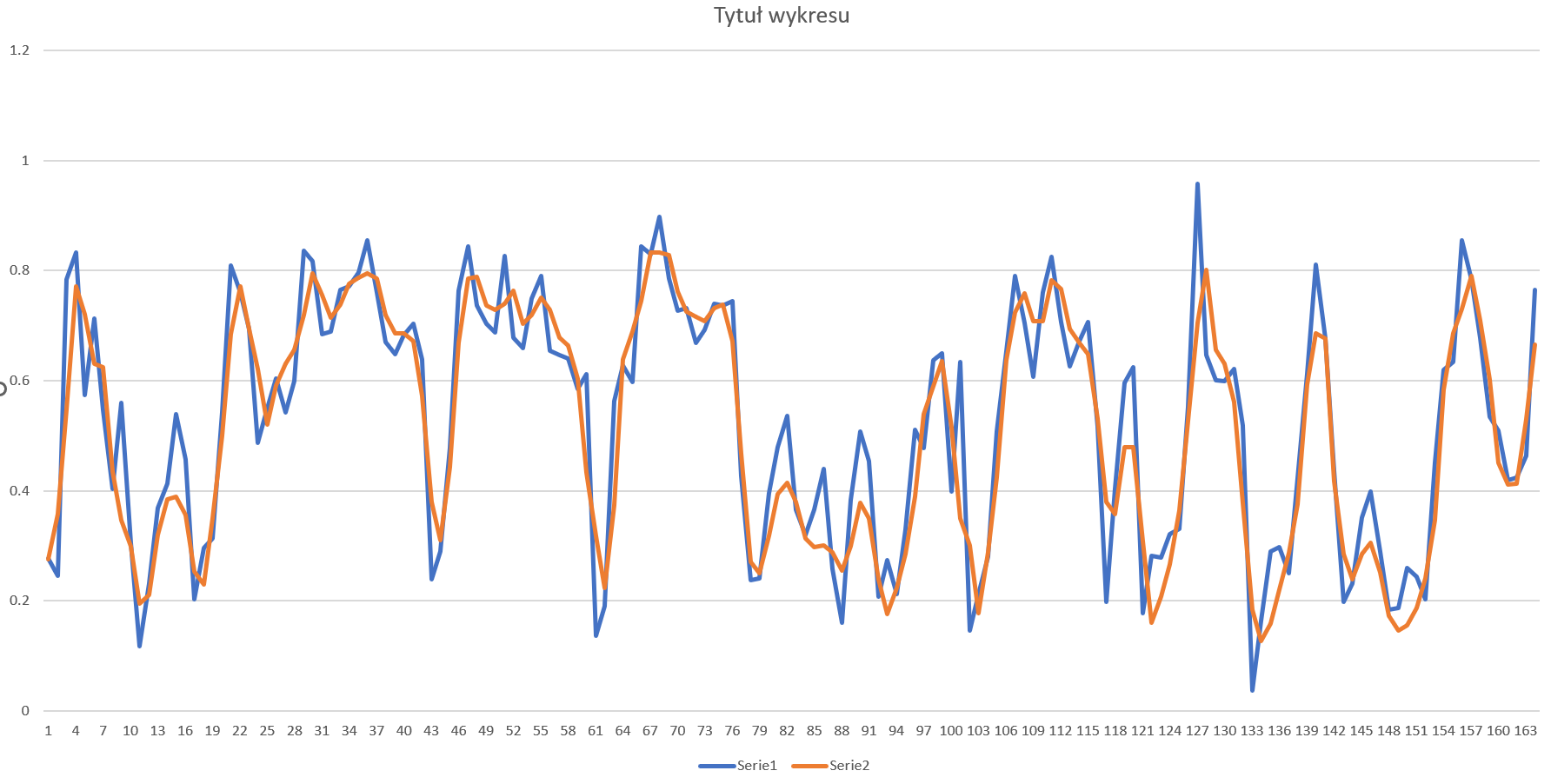
Can someone please elaborate why shuffling the data criplles the model? Or not shuffling the data in case of LSTM is just as bad as in case of other models?
I am trying to make an LSTM to predict the next value of an indicator but it predicts mean.
Data:
(Note: Data preparation function is on the bottom of the post so the post itself will be more readable)
I have around 25 000 entries in each data record and I have 14 columns of characteristics.
So my main array is 25 000 x 14.
When I prepare my data I am creating sequences in a shape of [number of sequences, samples in a sequence, features] and from then on 6 sets of data:
- X_train, Y_train
- X_valid, Y_valid
- X_test, Y_test
Where Y test is the one step ahead value of a feature I am trying to predict.
Note:
All datasets are scaled with MinMaxScaler in range (-1, 1) hence some data is below zero.
The value I am trying to predicts behaves in a following manner (previous values are inside X datasets):
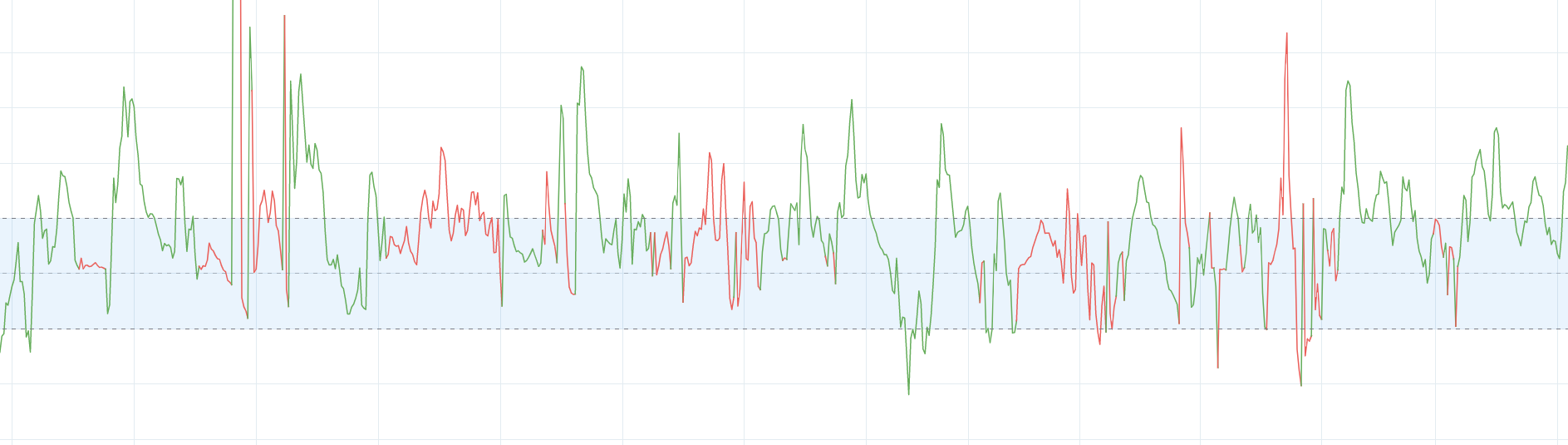
Example of the data sample: (Hence, different level of values I've plotted some series on another chart):
The Problem:
The problem is that no matter how many neurons, layers, what activation functions I use it predicts the mean value of a characteristic no matter what, and basically when the neural net hits loss of value around 0.078 the loss stops decreasing, If I waint longer and give it more epochs on the same learning rate sometimes loss skyrockets to 'NaN or 10^30.
Here is my Model:
X_train, Y_train, X_valid, Y_valid, X_test, Y_test, scaler = prepare_datasets_lstm_backup(dataset=dataset, samples=200)
optimizer = keras.optimizers.Adam(learning_rate=0.001)
initializer = keras.initializers.he_normal
model = keras.models.Sequential()
model.add(keras.layers.LSTM(64, activation='relu', input_shape=(200, 14), return_sequences=True))
model.add(keras.layers.LSTM(64, activation='relu', return_sequences=True))
model.add(keras.layers.LSTM(3, kernel_regularizer='l2', bias_regularizer='l2', return_sequences=False))
model.add(keras.layers.Activation('sigmoid'))
model.compile(loss='mse', optimizer=optimizer)
history = model.fit(X_train, Y_train, epochs=10, validation_data=(X_valid, Y_valid))
plt.plot(history.history['loss'], label='loss')
plt.plot(history.history['val_loss'], label='val_loss')
plt.legend()
plt.xlabel('Epochs')
plt.ylabel('loss function value')
plt.grid()
plt.show()
prediction = model.predict(X_test)
The possible solution
While simply increasing number of neurons and layers didn't helped I found a post on CrossValidated stackexchange forum: