I want to identify the clinics (csv file with longitude & latitude information and clinic ID) under the wildfire smoke polygon (shapefile) which is available at daily level from 2005 to 2019.
I could identify the clinics with a single wildfire smoke polygon with the R code below.
hms191030 <-readOGR("/Users/jin/Documents/HMS/hms_smoke20191030.shp")
clinics <-read_csv('/Users/jin/Documents/ESRD/Clinics_cont.csv')
clinicsW <-subset(clinics, PHYS_STATE_NM=="WASHINGTON" | PHYS_STATE_NM=="OREGON" | PHYS_STATE_NM=="CALIFORNIA")
clinicsW_sf <- st_as_sf(clinicsW, coords = c("LONG", "LAT"), crs = 4326)
hms191030_sf <- st_as_sf(hms191030, coords = c("long", "lat"), crs = 4326)
clinicsW_sf <- st_as_sf(clinicsW , coords = c("LONG", "LAT"), crs = 4326)
sf::sf_use_s2(FALSE)
intersection <- st_intersection(x = hms191030_sf, y = clinicsW_sf)
As I have to repeat this process for the multiple daily wildfire smoke polygons (365 day*15 years), I'm trying to use loop function with the subset of the shapefiles (from Jan 1 2019 to Jan 5 2019).
# 1. Open clinics data
clinics <-read_csv('/Users/songhyeonjin/Documents/ESRD/Clinics_cont.csv')
clinicsW <-subset(clinics, PHYS_STATE_NM=="WASHINGTON" | PHYS_STATE_NM=="OREGON" | PHYS_STATE_NM=="CALIFORNIA")
clinicsW_sf <- st_as_sf(clinicsW, coords = c("LONG", "LAT"), crs = 4326)
# 2. Open multiple HMS shapefiles and make a list
setwd("/Users/songhyeonjin/Documents/HMS/Shapefile_yrly/2019sub")
shps <- dir(getwd(), recursive = TRUE,"*.shp")
for (shp in shps) assign(shp, readOGR(shp))
my.list <-lapply(paste0('hms_smoke',20190101:20190105,'.shp'),get)
for (h in my.list) {
st_as_sf(h, coords = c("long", "lat"), crs = 4326)
}
# 3. Apply intersection function to the list of SpatialPolygonsDataFrame
sf::sf_use_s2(FALSE)
my_fun <- function(b) {
intersection <- st_intersection(x = b, y = clinicsW_sf)
return(intersection)
}
lapply(my.list, my_fun)
Then for the process #3, I get the error message
Error in UseMethod("st_intersection") :
no applicable method for 'st_intersection' applied to an object of class "c('SpatialPolygonsDataFrame', 'SpatialPolygons', 'Spatial', 'SpatialVector', 'SpatialPolygonsNULL')"
What I'm expecting to get is a data frame which shows the clinic ID that is under the wildfire smoke from 2005 to 2019.
It is my first time to ask a question in stack overflow. Thank you in advance and I really appreciate your help.
[**This is the updated code with example data]
# Example data for the clinics location
ID <- c(101, 102, 103, 104, 105)
LAT <- c(33.78595, 34.26310, 44.64489, 46.19070, 47.20550)
LONG <- c(-118.1900, -119.2293, -123.1116, -123.8365, -123.7543)
STATE <- c("California", "California", "Oregon", "Oregon", "Washington")
clinicsW <- data.frame(ID, LAT, LONG, STATE)
# Wildfire smoke shapefiles can be downlaoded here by selecting 'SMOKE' and 'Shapefile' and date range (2019 Jan 1 - 2019 Jan 5)
<https://www.ospo.noaa.gov/Products/land/hms.html#data>
# Edited code by John
setwd("/Users/songhyeonjin/Documents/HMS/Shapefile_yrly/2019sub")
shps <- dir(getwd(), recursive = TRUE,"*.shp")
# Just read in the shapefiles with sf to start.
# Then you don't need the conversion.
for (shp in shps) assign(shp, st_read(shp))
my.list <-lapply( paste0('hms_smoke',20190101:20190105,'.shp'),get)
# 3. Apply intersection function to the list of sf objects
sf::sf_use_s2(FALSE)
my_fun <- function(b) {
intersection <- st_intersection(x = b, y = clinicsW_sf)
return(intersection)
}
lapply(my.list, my_fun)
CodePudding user response:
I don't have your data, which makes this harder. You are starting with sp objects and converting some of them to sf objects, but not converting all of them. I think the problem is in your second step where you attempt to convert. I think you're not actually converting anything. You probably want something like this.
setwd("/Users/songhyeonjin/Documents/HMS/Shapefile_yrly/2019sub")
shps <- dir(getwd(), recursive = TRUE,"*.shp")
# Just read in the shapefiles with sf to start.
# Then you don't need the conversion.
for (shp in shps) assign(shp, st_read(shp))
my.list <-lapply( paste0('hms_smoke',20190101:20190105,'.shp'),get)
# 3. Apply intersection function to the list of sf objects
sf::sf_use_s2(FALSE)
my_fun <- function(b) {
intersection <- st_intersection(x = b, y = clinicsW_sf)
return(intersection)
}
lapply(my_list, my_fun)
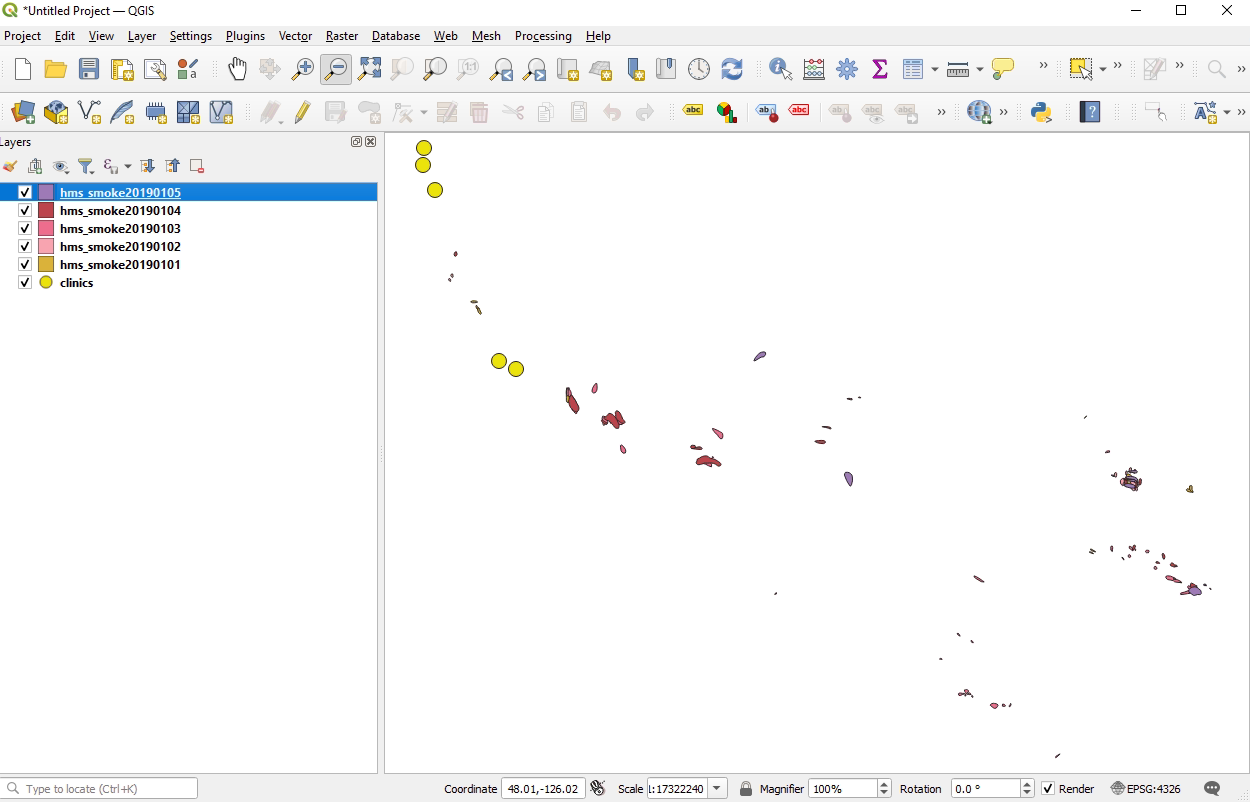
Here is a screenshot of the files in QGIS. Since the clinics are on the bottom of the Table of Contents there, any smoke shapefiles that would intersect would appear on top of the clinic points. None of them overlap, e.g. there is no intersection.
It's not that st_intersections doesn't work. With no intersections, there will be 0 rows returned.
At this point, all of the code works.
CodePudding user response:
So here is the updated code that works based on John's answer. Hope this is helpful to others as well!
# 1. Open clinics data
clinics <-read_csv('/Users/jin/Documents/ESRD/Clinics_cont.csv')
clinicsW <-subset(clinics, PHYS_STATE_NM=="WASHINGTON" | PHYS_STATE_NM=="OREGON" | PHYS_STATE_NM=="CALIFORNIA")
clinicsW_sf <- st_as_sf(clinicsW, coords = c("LONG", "LAT"), crs = 4326)
# 2. Open multiple HMS shapefiles and make a list
setwd("/Users/jin/Documents/HMS/Shapefile_yrly/2019sub")
shps <- dir(getwd(), recursive = TRUE,"*.shp")
for (shp in shps) assign(shp, readOGR(shp))
my.list <-lapply(paste0('hms_smoke',20190101:20190105,'.shp'),get)
for (h in my.list) {
st_as_sf(h, coords = c("long", "lat"), crs = 4326)
}
# 3. Apply intersection function to the list of SpatialPolygonsDataFrame
sf::sf_use_s2(FALSE)
newlist<-lapply(my.list, function(b) st_intersection(x=b, y=clinicsW_sf))