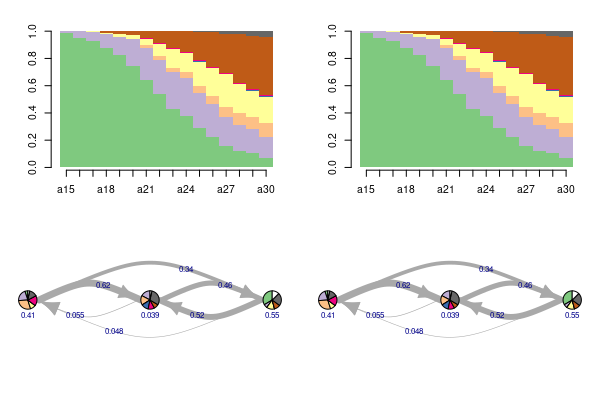
I want to plot two sequence distribution plots with the TraMineR package (seqplot(type = 'd')) and below them two Hidden Markov Models which correspond to the sequence plots (those are clusters).
For the reprex, I just use the build in biofam data set. Using layout() produces the desired result up to the third plot. The fourth plot is drawn to a new page, so essentially the bottom right space is left empty, but the first three panes are used accordingly to the layout() specs.
library('seqHMM')
library('TraMineR')
data("biofam")
biofam <- biofam[sample(nrow(biofam),300),]
biofam.lab <- c("Parent", "Left", "Married", "Left Marr",
"Child", "Left Child", "Left Marr Child", "Divorced")
biofam.seq <- seqdef(biofam, 10:25, labels=biofam.lab)
#> [>] 8 distinct states appear in the data:
#> 1 = 0
#> 2 = 1
#> 3 = 2
#> 4 = 3
#> 5 = 4
#> 6 = 5
#> 7 = 6
#> 8 = 7
#> [>] state coding:
#> [alphabet] [label] [long label]
#> 1 0 0 Parent
#> 2 1 1 Left
#> 3 2 2 Married
#> 4 3 3 Left Marr
#> 5 4 4 Child
#> 6 5 5 Left Child
#> 7 6 6 Left Marr Child
#> 8 7 7 Divorced
#> [>] 300 sequences in the data set
#> [>] min/max sequence length: 16/16
### Plot the sequence
seqplot(biofam.seq,type='d',with.legend=FALSE)
### Fit a Hidden Markov Model
biofam.hmm <- build_hmm(observations = biofam.seq, n_states = 3)
### Plot the HMM once
plot(biofam.hmm, with.legend=FALSE)
### Use layout to plot two sequence distribution plots and below there HMM
layout(matrix(c(1,2,3,4), ncol = 2, byrow = TRUE))
layout.show(4)
par(cex = 0.65, mar = c(2.5,4.1,2.5,1.5))
### The Sequence plots
seqplot(biofam.seq, type = 'd', with.legend = FALSE, border = NA, ylab = NA)
seqplot(biofam.seq, type = 'd', with.legend = FALSE, border = NA, ylab = NA)
### Change the par() arguments
par(cex = 0.5, mar = c(1.5,3.5,3.5,1.5))
plot(biofam.hmm, with.legend = FALSE, vertex.size = 15)
plot(biofam.hmm, with.legend = FALSE, vertex.size = 15)