Quick question here. I am trying to use the interactions package for R, to plot categorical interactions from a logistic regression model. Basically, let's say that I have a regression in which I have an interaction factor1*factor2, I would like to produce a plot with the OR and 95%CI of factor2 levels across factor1 levels, also reporting the p for interaction.
This is my current code:
library(interactions)
library(ggplot2)
library(dplyr)
set.seed(154)
outcome=sample(c(0,1), 1000, replace=TRUE)
set.seed(158)
factor1=sample(c("A","B"), 1000, replace=TRUE)
set.seed(1258)
factor2=sample(c("D","F"), 1000, replace=TRUE)
df <- data.frame(outcome, factor1, factor2)
df$outcome <- as.factor(df$outcome)
fit3 <- glm(outcome ~ factor1*factor2, data = df, family=binomial(link="logit"))
cat_plot(fit3, pred = factor1, modx = factor2, interval = TRUE)
Which produces the following outcome:
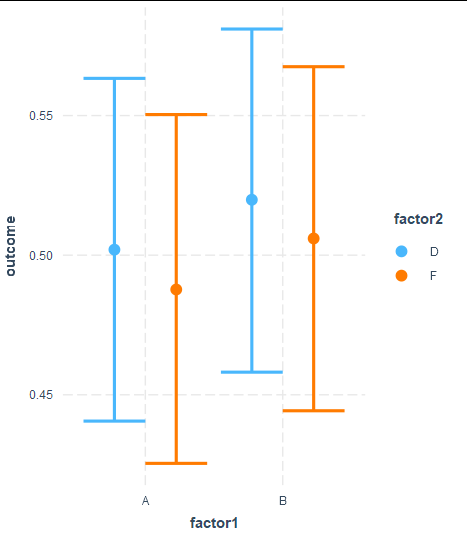
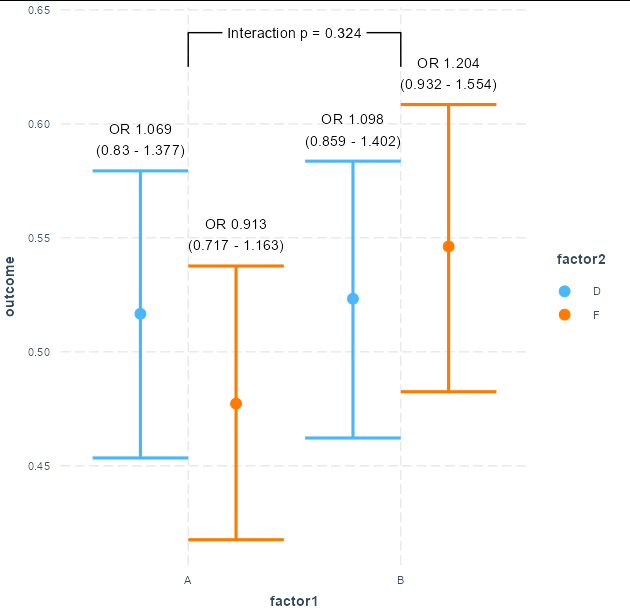
However, I was unable to add OR and 95%CI labels on top of each lines, and I was also unable to add p values for interaction. Is there a way I can do this?
CodePudding user response:
There is a trade-off with ggplot wrappers like cat_plot. What you gain in ease-of-use, you lose in ability to customize the plot. I would tend to use vanilla ggplot in these circumstances, but in this case it's not terribly difficult to add appropriate text labels by adding these on as layers to your existing cat_plot
library(geomtextpath)
p <- cat_plot(fit3, pred = factor1, modx = factor2, interval = TRUE)
p2o <- function(x) round(x/(1 - x), 3)
p
geom_textpath(data = data.frame(factor1 = c('A', 'A', 'B', 'B'),
factor2 = 'D',
outcome = c(0.625, 0.64, 0.64, 0.625)),
aes(label = paste('Interaction p =',
scales::pvalue(summary(fit3)$coef[4, 4]))),
color = 'black')
geom_text(aes(y = ymax, label = paste0("OR ", p2o(outcome), '\n(',
p2o(ymin), ' - ', p2o(ymax), ')'),
group = factor2),
color = 'black', position = position_dodge(width = 0.9),
vjust = -0.5)