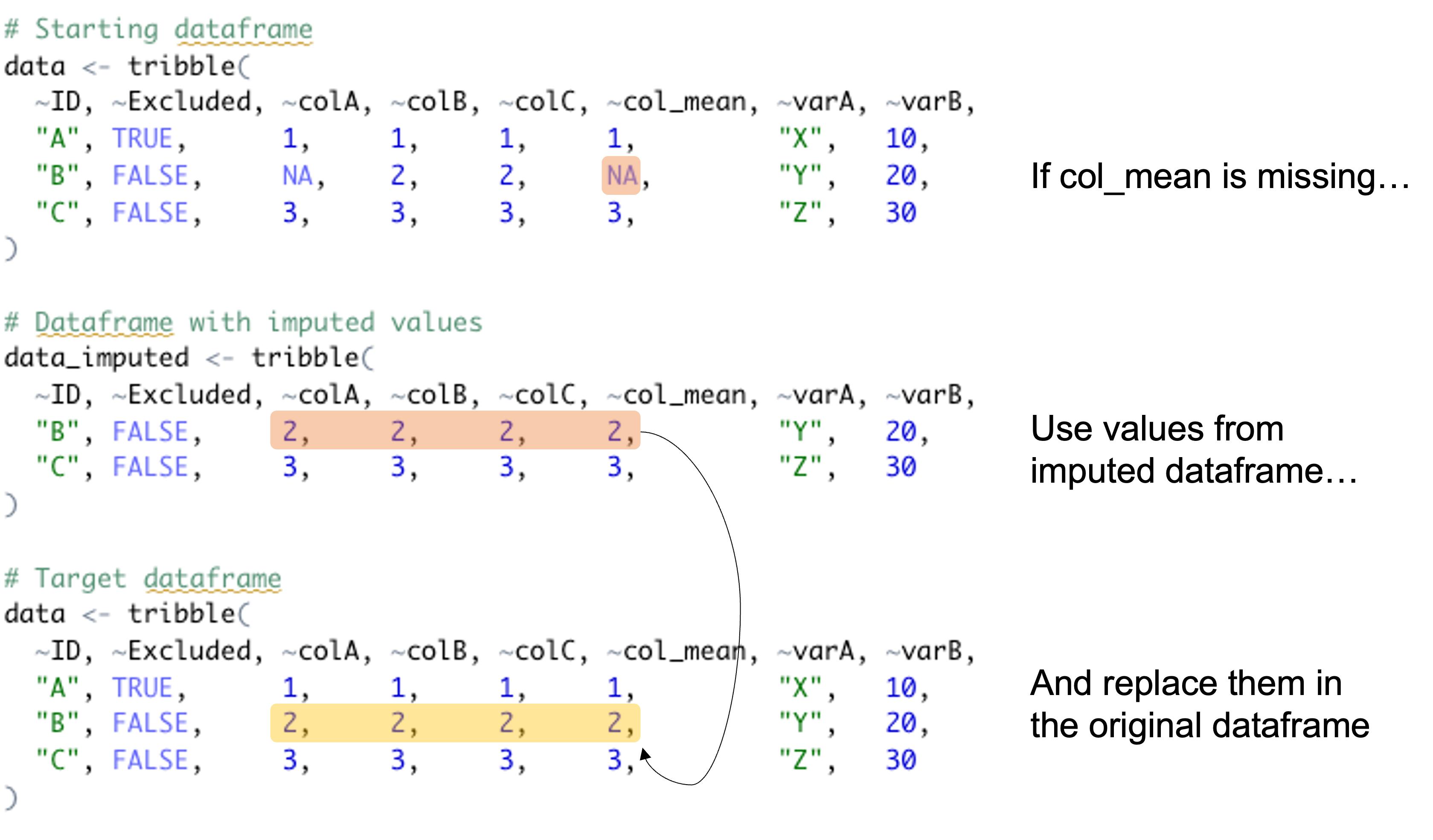
Suppose I have the following dataframe...
# Starting dataframe
data <- tribble(
~ID, ~Excluded, ~colA, ~colB, ~colC, ~col_mean, ~varA, ~varB,
"A", TRUE, 1, 1, 1, 1, "X", 10,
"B", FALSE, NA, 2, 2, NA, "Y", 20,
"C", FALSE, 3, 3, 3, 3, "Z", 30
)
And a subsetted dataframe (i.e. fewer observations exist) where missing values have been imputed, e.g. ...
# Dataframe with imputed values
data_imputed <- tribble(
~ID, ~Excluded, ~colA, ~colB, ~colC, ~col_mean, ~varA, ~varB,
"B", FALSE, 2, 2, 2, 2, "Y", 20,
"C", FALSE, 3, 3, 3, 3, "Z", 30
)
How do I replace values in the original dataframe with those from the imputed dataframe when a particular column (e.g. col_mean) has a missing value?
Note: I don't want to replace the whole row with the row from the imputed dataframe, just a specified set of columns (e.g., in this case, those starting with "col").
The target dataframe would look like this...
# Target dataframe
data <- tribble(
~ID, ~Excluded, ~colA, ~colB, ~colC, ~col_mean, ~varA, ~varB,
"A", TRUE, 1, 1, 1, 1, "X", 10,
"B", FALSE, 2, 2, 2, 2, "Y", 20,
"C", FALSE, 3, 3, 3, 3, "Z", 30
)
I have tried to summarise the problem with this figure...
I need to do this for four or five sets of columns, so something where I can specify the condition (e.g. is.na(col_mean)) and the columns to use (using regex) would make things easier.
I tend to use tidyverse, so code that works with tidyverse syntax is preferred.
CodePudding user response:
You could use the rows_ family from dplyr. In this case, rows_patch() works well. It modifies existing rows by some key columns (i.e. ID), but only overwrites NA values.
library(dplyr)
rows_patch(data, data_imputed, by = "ID")
# # A tibble: 3 × 8
# ID Excluded colA colB colC col_mean varA varB
# <chr> <lgl> <dbl> <dbl> <dbl> <dbl> <chr> <dbl>
# 1 A TRUE 1 1 1 1 X 10
# 2 B FALSE 2 2 2 2 Y 20
# 3 C FALSE 3 3 3 3 Z 30
CodePudding user response:
left_join(data, data_imputed, by = "ID") %>%
mutate(across(ends_with(".x"), ~ coalesce(., cur_data()[[sub("\\.x$", ".y", cur_column())]]))) %>%
select(-ends_with(".y")) %>%
rename_with(.fn = ~ sub("\\.x$", "", .))
# # A tibble: 3 x 8
# ID Excluded colA colB colC col_mean varA varB
# <chr> <lgl> <dbl> <dbl> <dbl> <dbl> <chr> <dbl>
# 1 A TRUE 1 1 1 1 X 10
# 2 B FALSE 2 2 2 2 Y 20
# 3 C FALSE 3 3 3 3 Z 30