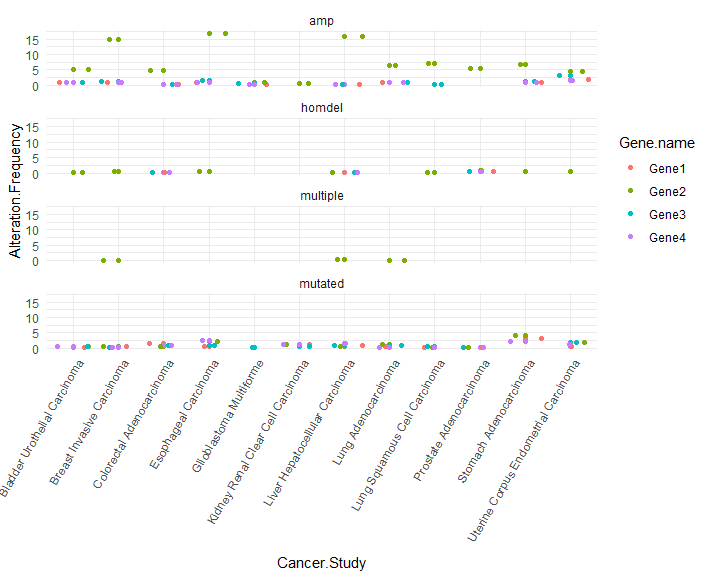
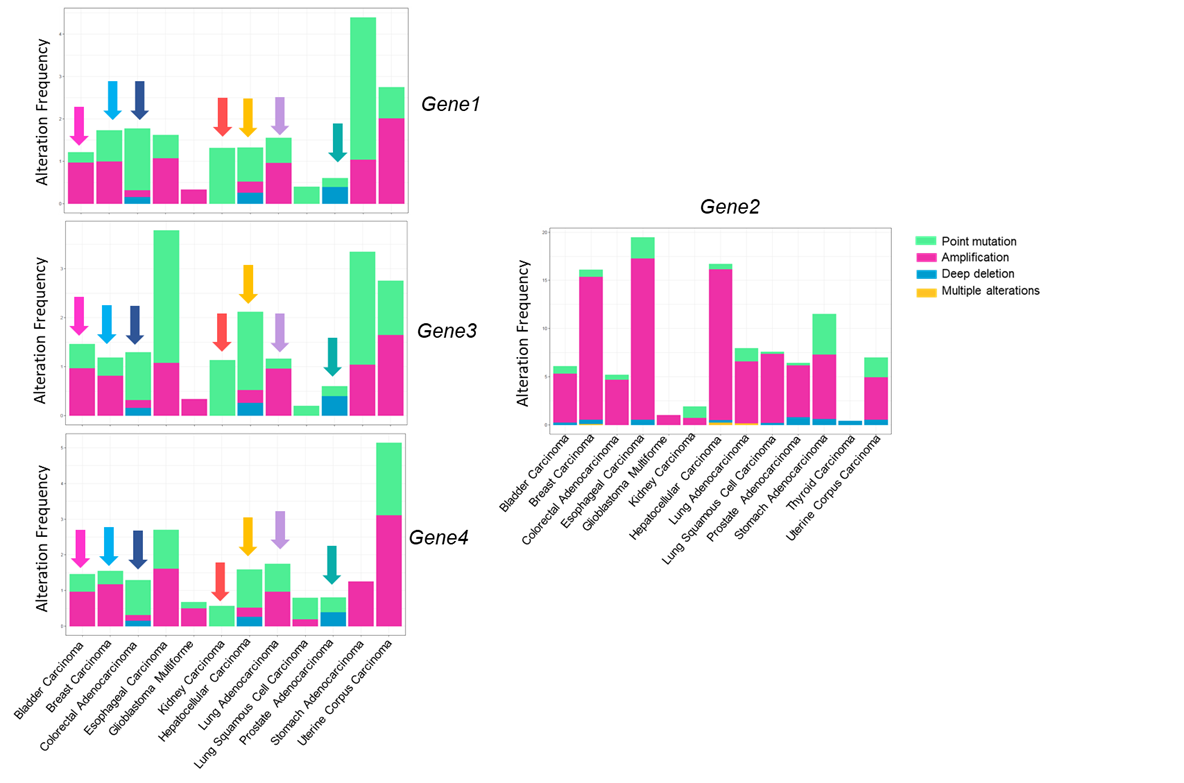
I have four barplots. Three of them have similar patterns but one behaves differently. How can I show this difference in R? As you can see I've used colored-coded arrows, but I like to quantify the similarity between the three plots and their difference from the fourth.
 Thanks for any help.
Here is my data:
Thanks for any help.
Here is my data:
dput(data)
structure(list(Gene.name = c("Gene1", "Gene1", "Gene1", "Gene1",
"Gene1", "Gene1", "Gene1", "Gene1", "Gene1", "Gene1", "Gene1",
"Gene1", "Gene1", "Gene1", "Gene1", "Gene1", "Gene1", "Gene1",
"Gene1", "Gene1", "Gene1", "Gene1", "Gene1", "Gene2", "Gene2",
"Gene2", "Gene2", "Gene2", "Gene2", "Gene2", "Gene2", "Gene2",
"Gene2", "Gene2", "Gene2", "Gene2", "Gene2", "Gene2", "Gene2",
"Gene2", "Gene2", "Gene2", "Gene2", "Gene2", "Gene2", "Gene2",
"Gene2", "Gene2", "Gene2", "Gene2", "Gene2", "Gene2", "Gene2",
"Gene2", "Gene2", "Gene2", "Gene2", "Gene3", "Gene3", "Gene3",
"Gene3", "Gene3", "Gene3", "Gene3", "Gene3", "Gene3", "Gene3",
"Gene3", "Gene3", "Gene3", "Gene3", "Gene3", "Gene3", "Gene3",
"Gene3", "Gene3", "Gene3", "Gene3", "Gene3", "Gene3", "Gene3",
"Gene4", "Gene4", "Gene4", "Gene4", "Gene4", "Gene4", "Gene4",
"Gene4", "Gene4", "Gene4", "Gene4", "Gene4", "Gene4", "Gene4",
"Gene4", "Gene4", "Gene4", "Gene4", "Gene4", "Gene4", "Gene4",
"Gene4", "Gene4"), Cancer.Study = c("Stomach Adenocarcinoma ",
"Stomach Adenocarcinoma ", "Uterine Corpus Endometrial Carcinoma ",
"Uterine Corpus Endometrial Carcinoma ", "Colorectal Adenocarcinoma ",
"Colorectal Adenocarcinoma ", "Colorectal Adenocarcinoma ", "Breast Invasive Carcinoma ",
"Breast Invasive Carcinoma ", "Esophageal Carcinoma ", "Esophageal Carcinoma ",
"Lung Adenocarcinoma ", "Lung Adenocarcinoma ", "Liver Hepatocellular Carcinoma ",
"Liver Hepatocellular Carcinoma ", "Liver Hepatocellular Carcinoma ",
"Kidney Renal Clear Cell Carcinoma ", "Bladder Urothelial Carcinoma ",
"Bladder Urothelial Carcinoma ", "Prostate Adenocarcinoma ",
"Prostate Adenocarcinoma ", "Lung Squamous Cell Carcinoma ",
"Glioblastoma Multiforme ", "Esophageal Carcinoma", "Esophageal Carcinoma",
"Esophageal Carcinoma", "Liver Hepatocellular Carcinoma", "Liver Hepatocellular Carcinoma",
"Liver Hepatocellular Carcinoma", "Liver Hepatocellular Carcinoma",
"Breast Invasive Carcinoma", "Breast Invasive Carcinoma", "Breast Invasive Carcinoma",
"Breast Invasive Carcinoma", "Stomach Adenocarcinoma", "Stomach Adenocarcinoma",
"Stomach Adenocarcinoma", "Lung Adenocarcinoma", "Lung Adenocarcinoma",
"Lung Adenocarcinoma", "Lung Squamous Cell Carcinoma", "Lung Squamous Cell Carcinoma",
"Lung Squamous Cell Carcinoma", "Uterine Corpus Endometrial Carcinoma",
"Uterine Corpus Endometrial Carcinoma", "Uterine Corpus Endometrial Carcinoma",
"Prostate Adenocarcinoma", "Prostate Adenocarcinoma", "Prostate Adenocarcinoma",
"Bladder Urothelial Carcinoma", "Bladder Urothelial Carcinoma",
"Bladder Urothelial Carcinoma", "Colorectal Adenocarcinoma",
"Colorectal Adenocarcinoma", "Kidney Renal Clear Cell Carcinoma",
"Kidney Renal Clear Cell Carcinoma", "Glioblastoma Multiforme",
"Uterine Corpus Endometrial Carcinoma ", "Uterine Corpus Endometrial Carcinoma ",
"Esophageal Carcinoma ", "Esophageal Carcinoma ", "Lung Adenocarcinoma ",
"Lung Adenocarcinoma ", "Liver Hepatocellular Carcinoma ", "Liver Hepatocellular Carcinoma ",
"Liver Hepatocellular Carcinoma ", "Breast Invasive Carcinoma ",
"Breast Invasive Carcinoma ", "Bladder Urothelial Carcinoma ",
"Bladder Urothelial Carcinoma ", "Colorectal Adenocarcinoma ",
"Colorectal Adenocarcinoma ", "Colorectal Adenocarcinoma ", "Stomach Adenocarcinoma ",
"Prostate Adenocarcinoma ", "Prostate Adenocarcinoma ", "Lung Squamous Cell Carcinoma ",
"Lung Squamous Cell Carcinoma ", "Glioblastoma Multiforme ",
"Glioblastoma Multiforme ", "Kidney Renal Clear Cell Carcinoma ",
"Esophageal Carcinoma ", "Esophageal Carcinoma ", "Stomach Adenocarcinoma ",
"Stomach Adenocarcinoma ", "Uterine Corpus Endometrial Carcinoma ",
"Uterine Corpus Endometrial Carcinoma ", "Liver Hepatocellular Carcinoma ",
"Liver Hepatocellular Carcinoma ", "Liver Hepatocellular Carcinoma ",
"Bladder Urothelial Carcinoma ", "Bladder Urothelial Carcinoma ",
"Colorectal Adenocarcinoma ", "Colorectal Adenocarcinoma ", "Colorectal Adenocarcinoma ",
"Breast Invasive Carcinoma ", "Breast Invasive Carcinoma ", "Lung Adenocarcinoma ",
"Lung Adenocarcinoma ", "Kidney Renal Clear Cell Carcinoma ",
"Prostate Adenocarcinoma ", "Prostate Adenocarcinoma ", "Glioblastoma Multiforme ",
"Lung Squamous Cell Carcinoma "), Alteration.Frequency = c(1.046025105,
3.347280335, 2.018348624, 0.733944954, 0.161550889, 0.161550889,
1.453957997, 1.000909918, 0.727934486, 1.081081081, 0.540540541,
0.968992248, 0.581395349, 0.265251989, 0.265251989, 0.795755968,
1.31826742, 0.97323601, 0.243309002, 0.400801603, 0.200400802,
0.399201597, 0.336700337, 0.540540541, 16.75675676, 2.162162162,
0.265251989, 0.265251989, 15.64986737, 0.530503979, 0.090991811,
0.454959054, 14.83166515, 0.727934486, 0.627615063, 6.694560669,
4.184100418, 0.19379845, 6.395348837, 1.356589147, 0.199600798,
7.185628743, 0.199600798, 0.550458716, 4.403669725, 2.018348624,
0.801603206, 5.410821643, 0.200400802, 0.243309002, 5.109489051,
0.729927007, 4.684975767, 0.484652666, 0.753295669, 1.129943503,
1.01010101, 3.119266055, 2.018348624, 1.621621622, 1.081081081,
0.968992248, 0.775193798, 0.265251989, 0.265251989, 1.061007958,
1.18289354, 0.363967243, 0.97323601, 0.486618005, 0.161550889,
0.161550889, 0.969305331, 1.255230126, 0.400801603, 0.400801603,
0.199600798, 0.598802395, 0.505050505, 0.168350168, 0.564971751,
1.081081081, 2.702702703, 1.046025105, 2.30125523, 1.651376147,
1.100917431, 0.265251989, 0.265251989, 1.591511936, 0.97323601,
0.486618005, 0.161550889, 0.161550889, 0.969305331, 0.818926297,
0.363967243, 0.968992248, 0.19379845, 1.129943503, 0.400801603,
0.200400802, 0.336700337, 0.199600798), Alteration.Type = c("amp",
"mutated", "amp", "mutated", "homdel", "amp", "mutated", "amp",
"mutated", "amp", "mutated", "amp", "mutated", "homdel", "amp",
"mutated", "mutated", "amp", "mutated", "homdel", "mutated",
"mutated", "amp", "homdel", "amp", "mutated", "multiple", "homdel",
"amp", "mutated", "multiple", "homdel", "amp", "mutated", "homdel",
"amp", "mutated", "multiple", "amp", "mutated", "homdel", "amp",
"mutated", "homdel", "amp", "mutated", "homdel", "amp", "mutated",
"homdel", "amp", "mutated", "amp", "mutated", "amp", "mutated",
"amp", "amp", "mutated", "amp", "mutated", "amp", "mutated",
"homdel", "amp", "mutated", "amp", "mutated", "amp", "mutated",
"homdel", "amp", "mutated", "amp", "homdel", "mutated", "amp",
"mutated", "amp", "mutated", "mutated", "amp", "mutated", "amp",
"mutated", "amp", "mutated", "homdel", "amp", "mutated", "amp",
"mutated", "homdel", "amp", "mutated", "amp", "mutated", "amp",
"mutated", "mutated", "homdel", "mutated", "amp", "mutated")), class = "data.frame", row.names = c(NA,
-104L))
CodePudding user response:
If you want to still use a graph, but one where the comparison is easier, you could try separating the alteration types (edit: you have some trailing white spaces in your cancer study column which creates seemingly distinct categories)
library(ggplot2)
df$Cancer.Study=trimws(df$Cancer.Study)
ggplot(df,aes(y=Alteration.Frequency,x=Cancer.Study,color=Gene.name))
geom_point() geom_jitter()
facet_wrap(~Alteration.Type,ncol=1) theme_minimal()
theme(axis.text.x=element_text(angle=60,hjust=1))
CodePudding user response:
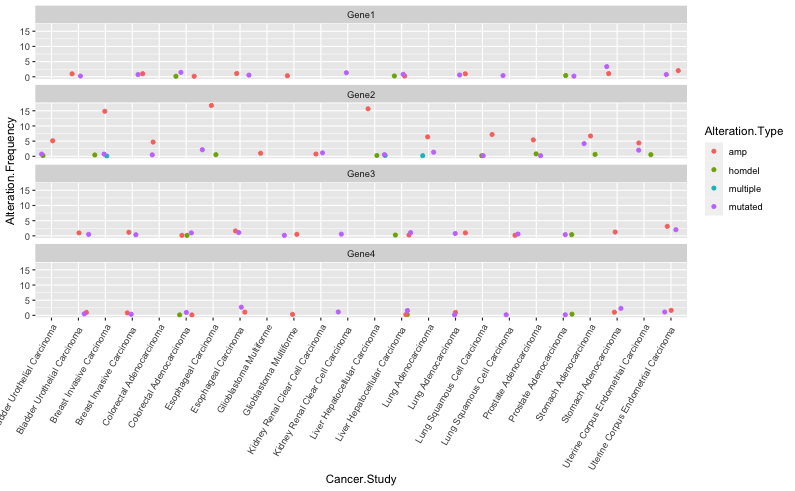
Configuring the faceted graph above like this could also help highlight the effect on "Gene2" with Alteration type "amp":
cancer_test <- cancer %>%
group_by(Cancer.Study)
ggplot(cancer_test, mapping = aes(x = Cancer.Study, y = Alteration.Frequency, color = Alteration.Type)) geom_jitter() facet_wrap(~Gene.name, ncol=1) theme(axis.text.x=element_text(angle=60, hjust=1))
CodePudding user response:
A numerical comparison can be done with dissimilarity indices like Chisquare or Bray-Curtis. Here we first need to convert the tidy data format (that is of course a good idea) into a cross table. Then we can apply the standard dist function or vegdistfrom package vegan.
The following approach uses pivot_wider from tidyr for the crosstable and then vegdist for the distance matrix. As the data types from tidyverse are not 100% compatible to vegan, we may convert it to a standard data.frame and then assign the names.
library(dplyr)
library(tidyr)
library(vegan)
## re-arrange as crosstable
crosstable <-
df %>%
pivot_wider(id_cols = c(Alteration.Type, Gene.name),
names_from = Cancer.Study,
values_from = Alteration.Frequency)
## show the result
crosstable
## convert to standard data frame, because tibbles don't support row names
crosstable <- as.data.frame(crosstable)
## assign row names and remove the ID columns
rownames(crosstable) <- with(crosstable, paste(Alteration.Type, Gene.name))
crosstable <- crosstable[,-c(1, 2)]
vegdist(crosstable, na.rm=TRUE, method="bray") # this is the default in vegan
## ==> we see lots of missing combinations,
## so one may consider further aggregation,
## e.g. combinations (means, sums, ...) of studies
## another possible dissimilarity measure if there were no missing data
# vegdist(crosstable, na.rm=TRUE, method="chisq")