I have a data (user_data) that represent the number of examples in each class (here we have 5 classes), for example in first row, 16 represent 16 samples in class 1 for user1, 15 represent that there is 15 samples belong to class 2 for user 1, ect.
user_data = np.array([
[16, 15, 14, 10, 0],
[0, 13, 6, 15, 21],
[12, 29, 1, 12, 1],
[0, 0, 0, 0, 55]])
I calculated the probability vector of each users by using method of Peter Flash’s book
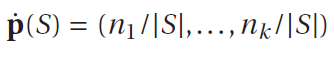
where |S|=55 and the number of examples in S of class Ci is denoted ni.
Output:
array([[0.29090909, 0.27272727, 0.25454545, 0.18181818, 0. ],
[0. , 0.23636364, 0.10909091, 0.27272727, 0.38181818],
[0.21818182, 0.52727273, 0.01818182, 0.21818182, 0.01818182],
[0. , 0. , 0. , 0. , 1. ]])
I used the following method to smooth all these frequencies to avoid issues with extreme values (0 or 1) by using Laplace smoothing where k=2.
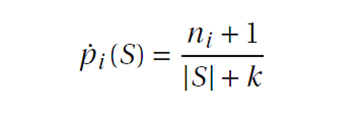
Output:
array([[0.29824561, 0.28070175, 0.26315789, 0.19298246, 0.01754386],
[0.01754386, 0.24561404, 0.12280702, 0.28070175, 0.38596491],
[0.22807018, 0.52631579, 0.03508772, 0.22807018, 0.03508772],
[0.01754386, 0.01754386, 0.01754386, 0.01754386, 0.98245614]])
But I want to smooth only extreme values (0 or 1) in this data, red Square represent extreme value:

So, only extreme values (0 and 1) are smoothed
CodePudding user response:
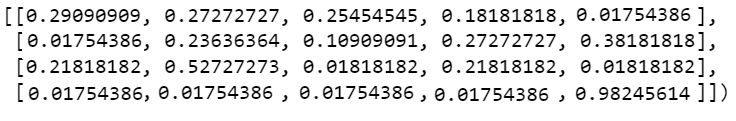
I have noticed that your smoothing approach will cause P > 1, you are to clip or normalize the values later on:
probs = user_data/55
alpha = (user_data 1)/(55 2)
extreme_values_mask = (probs == 0) | (probs == 1)
probs[extreme_values_mask] = alpha[extreme_values_mask]
Result:
array([[0.29090909, 0.27272727, 0.25454545, 0.18181818, 0.01754386],
[0.01754386, 0.23636364, 0.10909091, 0.27272727, 0.38181818],
[0.21818182, 0.52727273, 0.01818182, 0.21818182, 0.01818182],
[0.01754386, 0.01754386, 0.01754386, 0.01754386, 0.98245614]])
Extension:
# Scale by sum.
probs /= probs.sum(1).reshape((-1, 1))
print(probs)
print(probs.sum(1))
[[0.28589342 0.26802508 0.25015674 0.17868339 0.01724138]
[0.01724138 0.2322884 0.10721003 0.26802508 0.37523511]
[0.21818182 0.52727273 0.01818182 0.21818182 0.01818182]
[0.01666667 0.01666667 0.01666667 0.01666667 0.93333333]]
[1. 1. 1. 1.]
CodePudding user response:
You can use np.where, which follows the format np.where(condition, value if condition is true, value if condition is false) – feel free to read the documentation here.
For example:
S = 55
k = 2
np.where((user_data/S == 0) | (user_data/S == 1), (user_data 1)/(S k), user_data/S)
Result:
array([[0.29090909, 0.27272727, 0.25454545, 0.18181818, 0.01754386],
[0.01754386, 0.23636364, 0.10909091, 0.27272727, 0.38181818],
[0.21818182, 0.52727273, 0.01818182, 0.21818182, 0.01818182],
[0.01754386, 0.01754386, 0.01754386, 0.01754386, 0.98245614]])