I am plotting two lists of data against each other, namely freq and data. Freq stands for frequency, and data are the numeric observations for each frequency.
In the next step, I apply the ordinary linear least-squared regression between freq and data, using stats.linregress on the logarithmic scale. My aim is applying the linear regression inside the log-log scale, not on the normal scale.
Before doing so, I transform both freq and data into np.log10, since I plan to plot a straight linear regression line on the logarithmic scale, using plt.loglog.
Problem:
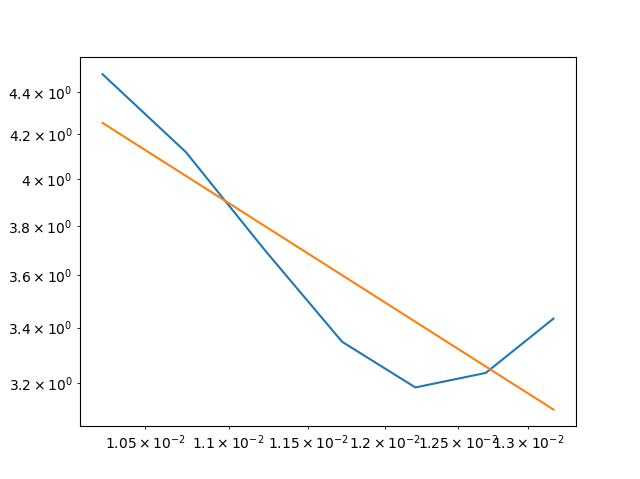
The problem is that the regression line, plotted in red color, is plotted far from the actual data, plotted in green color. I assume that there is a problem in combination with plt.loglog in my code, hence the visual distance between the green data and the red regression line. How can I fix this problem, so that the regression line plots on top of the actual data?
Here is my reproducible code:
import numpy as np
import matplotlib.pyplot as plt
from scipy import stats
# Data
freq = [0.0102539, 0.0107422, 0.0112305, 0.0117188, 0.012207, 0.0126953,
0.0131836]
data = [4.48575, 4.11893, 3.69591, 3.34766, 3.18452, 3.23554, 3.43357]
# Plot log10 of freq vs. data
plt.loglog(freq, data, c="green")
# Linear regression
log_freq = np.log10(freq)
log_data = np.log10(data)
reg = stats.linregress(log_freq, log_data)
slope = reg[0]
intercept = reg[1]
plt.plot(freq, slope*log_freq intercept, color="red")
And here is a screenshot of the code’s result:
CodePudding user response:
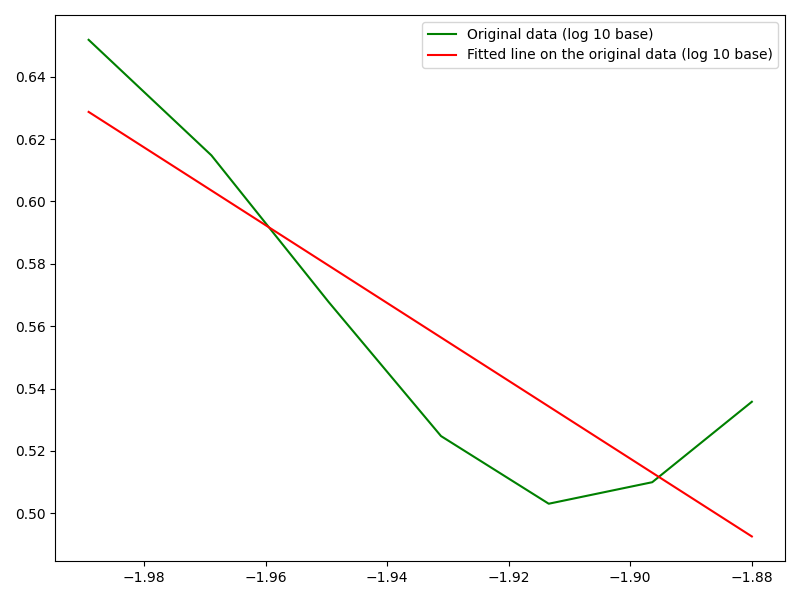
You can convert your data sets to log base 10 first, then do linear regression and plot them accordingly.
Note that after the log transformation, the numbers inlog_freq will all be negative; therefore x-axis cannot be log-scaled.
import numpy as np
import matplotlib.pyplot as plt
from scipy import stats
# Data
freq = np.array([0.0102539, 0.0107422, 0.0112305, 0.0117188, 0.012207, 0.0126953,
0.0131836])
data = np.array([4.48575, 4.11893, 3.69591, 3.34766, 3.18452, 3.23554, 3.43357])
# transform date to log base 10
log_freq = np.log10(freq)
log_data = np.log10(data)
# Plot freq vs. data
fig, ax = plt.subplots(figsize=(8, 6))
ax.plot(log_freq, log_data, c="green", label='Original data (log 10 base)')
# Linear regression
reg = stats.linregress(log_freq, log_data)
# Plot fitted freq vs. data
ax.plot(log_freq, reg.slope * log_freq reg.intercept, color="red",
label='Fitted line on the original data (log 10 base)')
plt.legend()
plt.tight_layout()
plt.show()
References:
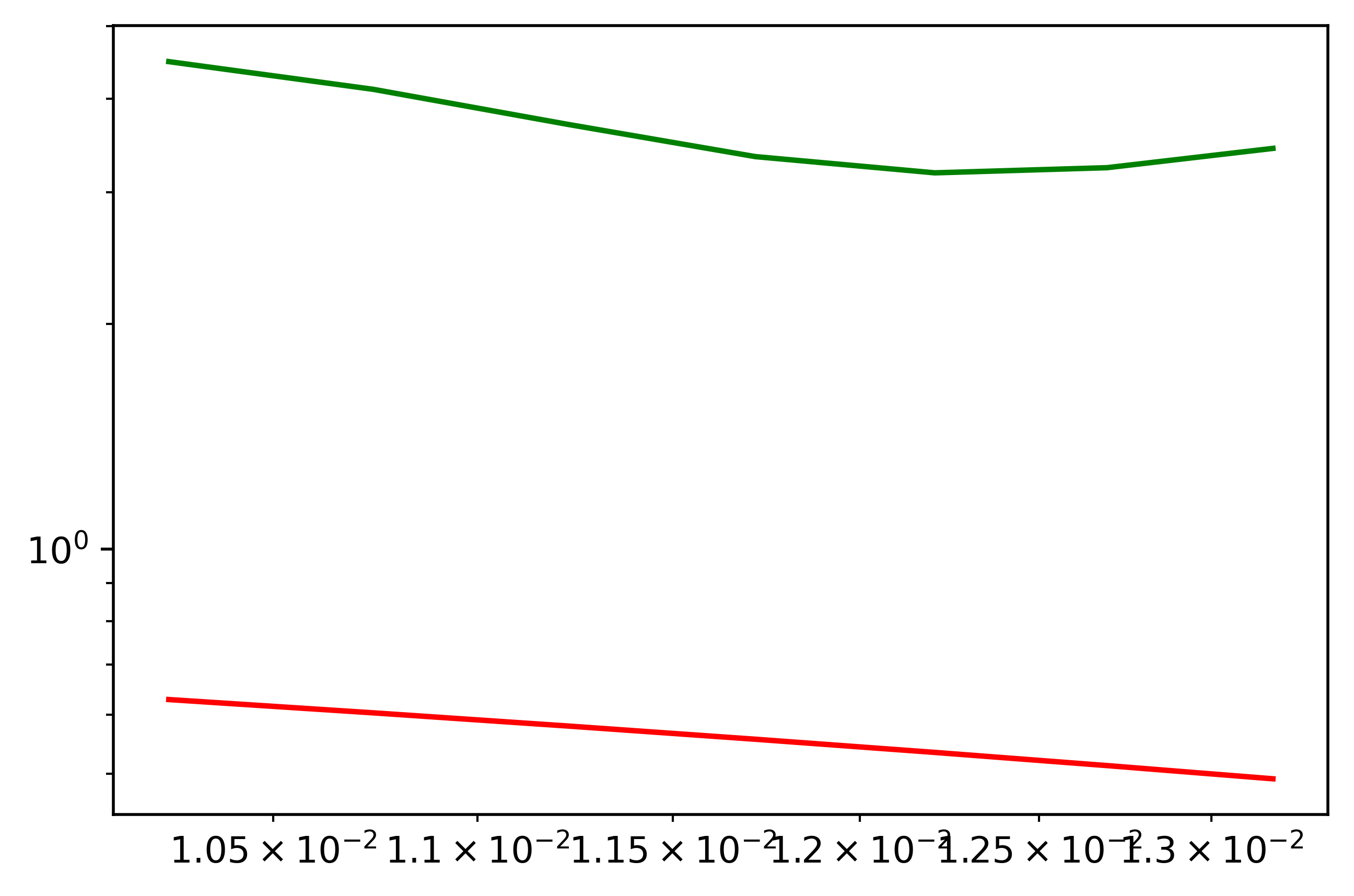
First of all, I question the necessity of log-log axes, because the ranges of the data, or at least the ranges of the data that you've shown us, are limited on both coordinates.
In the code below, I have
- computed the logarithms in base 10 of your arrays,
- used the formulas for linear regression but using the logarithms of data to obtain the equation of a straight line:
y = a b·x
in, so to say, the logarithmic space.
Because a straight line in log-space corresponds, in data-space, to a power law, y = pow(10, a)·pow(x, b), I have plotted
- the original data, in log-log, and
- the power law, also in log-log,
obtaining a straight line in the log-log representation.
import matplotlib.pyplot as plt
from math import log10
freq = [.0102539, .0107422, .0112305, .0117188, .012207, .0126953, .0131836]
data = [4.48575, 4.11893, 3.69591, 3.34766, 3.18452, 3.23554, 3.43357]
n = len(x)
lx, ly = [[log10(V) for V in v] for v in (freq, data)]
sum_x = sum(x for x in lx)
sum_y = sum(y for y in ly)
sum_x2 = sum(x**2 for x in lx)
sum_y2 = sum(y**2 for y in ly)
sum_xy = sum(x*y for x, y in zip(lx, ly))
# coefficients of a straight line "y = a b x" in log-log space
b = (n*sum_xy - sum_x*sum_y)/(n*sum_x2-sum_x**2)
a = (sum_y - b*sum_x)/n
plt.loglog(freq, data)
A = pow(10, a)
plt.loglog(freq, [A*pow(x, b) for x in freq])