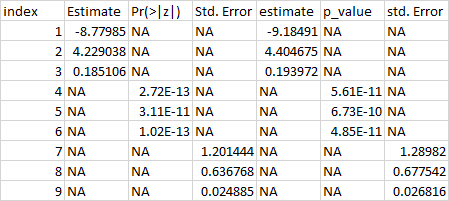
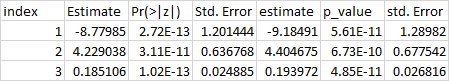
I have a different problem that I have not encountered yet. When I try to make a completely reproducible example, I get errors, so I am posting what my data look like, and what I would like it to look like. I am asking for suggestions on how to get the data into the shape I need to do some analyses.
Here is what the data look like:
If I am breaking some type of protocol, I apologize. When I try to make a reproducible example, I get the following error: "Error in [<-.data.frame(*tmp*, 2, 3, value = 3) :
new columns would leave holes after existing columns" This is the best I can do for some help.
I have tried binding rows, deleting rows with NAs, na.omit(), and several others. I just need to get the data to where it is no longer 7 columns and 9 rows, but to 7 columns and 3 rows. Thank you
CodePudding user response:
In base R you could do
as.data.frame(lapply(df1, na.omit))
For example:
df1 <- data.frame(a = c(1, 2, 3, NA, NA, NA, NA, NA, NA),
b = c(NA, NA, NA, 4, 5, 6, NA, NA, NA),
c = c(NA, NA, NA, NA, NA, NA, 7, 8, 9))
df1
#> a b c
#> 1 1 NA NA
#> 2 2 NA NA
#> 3 3 NA NA
#> 4 NA 4 NA
#> 5 NA 5 NA
#> 6 NA 6 NA
#> 7 NA NA 7
#> 8 NA NA 8
#> 9 NA NA 9
as.data.frame(lapply(df1, na.omit))
#> a b c
#> 1 1 4 7
#> 2 2 5 8
#> 3 3 6 9
Created on 2023-01-03 with reprex v2.0.2
CodePudding user response:
We may loop across columns other than index, order based on the NA elements so that all non-NAs order at the top followed by NA and then use filter to remove the rows having all NAs with if_any
library(dplyr)
df1 %>%
mutate(across(-index, ~ .x[order(is.na(.x))])) %>%
filter(if_any(-index, complete.cases))
NOTE: This will keep some NA rows in case there are difference in number of NAs per each columns