I have a dataframe with a row full of adverse events but also relationships of these adverse events to the procedure, like this:
df <- data.frame(
adverse_event = c(
"Haemorrhage", "related", "likely related",
"Other", "related", "likely related", "Pain", "related", "likely related",
"Subcapsular hematoma", "related", "likely related", "Ascites",
"related", "likely related", "Hyperbilirubinemia", "related",
"likely related", "Liver abscess", "related", "likely related",
"Pleural effusion with drainage", "related", "likely related",
"Pneumothorax", "related", "likely related", "Biliary leakage / occlusion / fistula",
"related", "likely related", "Portal vein thrombosis", "related",
"likely related", "Sepsis", "related", "likely related"
),
grade_1 = c(
4L, 4L, 0L, 3L, 6L, 1L, 8L, 4L, 5L, 3L, 1L, 3L, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA
),
grade_2 = c(
2L, 3L, 0L, 11L, 3L, 7L, 2L, 4L, 2L, 1L, 2L, 0L, 1L, 1L, 0L,
1L, 0L, 2L, 1L, 1L, 0L, 1L, 2L, 1L, 1L, 1L, 0L, NA, NA, NA, NA,
NA, NA, NA, NA, NA
),
grade_3 = c(
1L, 4L, 1L, 5L, 3L, 2L, 2L, 5L, 1L, NA, NA, NA, NA, NA, NA,
NA, NA, NA, 4L, 5L, 1L, NA, NA, NA, 1L, 1L, 0L, 1L, 2L, 0L, 1L,
1L, 0L, 1L, 1L, 0L
),
grade_4 = c(
2L, 4L, 1L, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA
)
)
Now I'd like to sort the adverse events alphabetically but of course take the "related", "likely related" rows with the individual adverse event rows, so I'd like to somehow group them first. In this example it's always 3 rows, but let's assume it could be sometimes 2, 4 or 5 rows too (all except the adverse event rows containing "related" in the string/name though e.g. 'unlikely related').
I know, I can get the indices of the adverse event rows by
grep('related', df$adverse_event, invert = T) but I'm unsure how to use this to group the rows together before sorting them.
Edit: Beginning of the left column of the desired output:
expected_output_left_column <- data.frame(adverse_event = c(
"Ascites", "related", "likely related",
"Biliary leakage / occlusion / fistula", "related", "likely related" ) )
Thank you!
CodePudding user response:
You can do the following:
library(dplyr)
left_join(
df,
df %>%
filter(!grepl('related',adverse_event)) %>%
select(adverse_event) %>%
arrange(adverse_event) %>%
mutate(o = row_number())
) %>%
mutate(o = data.table::nafill(o, "locf")) %>%
arrange(o) %>%
select(-o)
Output:
adverse_event grade_1 grade_2 grade_3 grade_4
1 Ascites NA 1 NA NA
2 related NA 1 NA NA
3 likely related NA 0 NA NA
4 Biliary leakage / occlusion / fistula NA NA 1 NA
5 related NA NA 2 NA
6 likely related NA NA 0 NA
7 Haemorrhage 4 2 1 2
8 related 4 3 4 4
9 likely related 0 0 1 1
10 Hyperbilirubinemia NA 1 NA NA
11 related NA 0 NA NA
12 likely related NA 2 NA NA
13 Liver abscess NA 1 4 NA
14 related NA 1 5 NA
15 likely related NA 0 1 NA
16 Other 3 11 5 NA
17 related 6 3 3 NA
18 likely related 1 7 2 NA
19 Pain 8 2 2 NA
20 related 4 4 5 NA
21 likely related 5 2 1 NA
22 Pleural effusion with drainage NA 1 NA NA
23 related NA 2 NA NA
24 likely related NA 1 NA NA
25 Pneumothorax NA 1 1 NA
26 related NA 1 1 NA
27 likely related NA 0 0 NA
28 Portal vein thrombosis NA NA 1 NA
29 related NA NA 1 NA
30 likely related NA NA 0 NA
31 Sepsis NA NA 1 NA
32 related NA NA 1 NA
33 likely related NA NA 0 NA
34 Subcapsular hematoma 3 1 NA NA
35 related 1 2 NA NA
36 likely related 3 0 NA NA
Note that this uses data.table::nafill().. A full data.table solution is as below:
library(data.table)
setDT(df)
data.table(adverse_event = sort(df[!grepl('related',adverse_event), adverse_event]))[, o:=.I][
df, on="adverse_event"][, o:=nafill(o, "locf")][order(o), !c("o")]
CodePudding user response:
Another solution using base r and lead function from dplyr
# where start each group
id <- grep('related', df$adverse_event, invert = T)
# size of each group
size <- lead(id) - id
size_of_last_group <- nrow(df) - id[length(id)] 1
size[length(size)] <- size_of_last_group
# add col with id
df$id <- paste0(rep(df$adverse_event[id], times = size),
df$adverse_event)
# order
df <- df[order(df$id), ]
# remove id
df$id <- NULL
CodePudding user response:
Just to throw in another tidyverse solution:
library(tidyr)
library(dplyr)
df %>%
mutate(grp = if_else(grepl("related", adverse_event),
NA_character_,
adverse_event)) %>%
fill(grp) %>%
nest(data = -grp) %>%
arrange(grp) %>%
unnest(cols = data) %>%
select(-grp)
# # A tibble: 36 × 5
# adverse_event grade_1 grade_2 grade_3 grade_4
# <chr> <int> <int> <int> <int>
# 1 Ascites NA 1 NA NA
# 2 related NA 1 NA NA
# 3 likely related NA 0 NA NA
# 4 Biliary leakage / occlusion / fistula NA NA 1 NA
# 5 related NA NA 2 NA
# 6 likely related NA NA 0 NA
# 7 Haemorrhage 4 2 1 2
# 8 related 4 3 4 4
# 9 likely related 0 0 1 1
# 10 Hyperbilirubinemia NA 1 NA NA
# 11 related NA 0 NA NA
# 12 likely related NA 2 NA NA
# 13 Liver abscess NA 1 4 NA
# 14 related NA 1 5 NA
# 15 likely related NA 0 1 NA
# 16 Other 3 11 5 NA
# 17 related 6 3 3 NA
# 18 likely related 1 7 2 NA
# 19 Pain 8 2 2 NA
# 20 related 4 4 5 NA
# 21 likely related 5 2 1 NA
# 22 Pleural effusion with drainage NA 1 NA NA
# 23 related NA 2 NA NA
# 24 likely related NA 1 NA NA
# 25 Pneumothorax NA 1 1 NA
# 26 related NA 1 1 NA
# 27 likely related NA 0 0 NA
# 28 Portal vein thrombosis NA NA 1 NA
# 29 related NA NA 1 NA
# 30 likely related NA NA 0 NA
# 31 Sepsis NA NA 1 NA
# 32 related NA NA 1 NA
# 33 likely related NA NA 0 NA
# 34 Subcapsular hematoma 3 1 NA NA
# 35 related 1 2 NA NA
# 36 likely related 3 0 NA NA
Explanation
mutate fill: Label eachadverse_eventwith the stem, i.e. re-label allrelatedrecords with the corresponding event above.- Nest all columns, but keep the newly created
grpcolumn, which bears the name of the stem adverse event. - Sort the adverse event stems.
- Unnest the rows again.
- Remove the
grpcolumn.
CodePudding user response:
An approach using rank. Using an extended data set with 4 entries for "Ascites".
library(dplyr)
df %>%
mutate(ord = !grepl("related", adverse_event),
grp = cumsum(ord),
Rank = rank(adverse_event[ord])[grp]) %>%
arrange(Rank) %>%
select(-c(ord, grp, Rank))
adverse_event grade_1 grade_2 grade_3 grade_4
1 Ascites NA 1 NA NA
2 related NA 1 NA NA
3 related NA 1 NA NA
4 likely related NA 0 NA NA
5 Biliary leakage / occlusion / fistula NA NA 1 NA
6 related NA NA 2 NA
7 likely related NA NA 0 NA
8 Haemorrhage 4 2 1 2
9 related 4 3 4 4
10 likely related 0 0 1 1
11 Hyperbilirubinemia NA 1 NA NA
12 related NA 0 NA NA
13 likely related NA 2 NA NA
14 Liver abscess NA 1 4 NA
15 related NA 1 5 NA
16 likely related NA 0 1 NA
17 Other 3 11 5 NA
18 related 6 3 3 NA
19 likely related 1 7 2 NA
20 Pain 8 2 2 NA
21 related 4 4 5 NA
22 likely related 5 2 1 NA
23 Pleural effusion with drainage NA 1 NA NA
24 related NA 2 NA NA
25 likely related NA 1 NA NA
26 Pneumothorax NA 1 1 NA
27 related NA 1 1 NA
28 likely related NA 0 0 NA
29 Portal vein thrombosis NA NA 1 NA
30 related NA NA 1 NA
31 likely related NA NA 0 NA
32 Sepsis NA NA 1 NA
33 related NA NA 1 NA
34 likely related NA NA 0 NA
35 Subcapsular hematoma 3 1 NA NA
36 related 1 2 NA NA
37 likely related 3 0 NA NA
extended data
df <- structure(list(adverse_event = c("Haemorrhage", "related", "likely related",
"Other", "related", "likely related", "Pain", "related", "likely related",
"Subcapsular hematoma", "related", "likely related", "Ascites",
"related", "related", "likely related", "Hyperbilirubinemia",
"related", "likely related", "Liver abscess", "related", "likely related",
"Pleural effusion with drainage", "related", "likely related",
"Pneumothorax", "related", "likely related", "Biliary leakage / occlusion / fistula",
"related", "likely related", "Portal vein thrombosis", "related",
"likely related", "Sepsis", "related", "likely related"), grade_1 = c(4L,
4L, 0L, 3L, 6L, 1L, 8L, 4L, 5L, 3L, 1L, 3L, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA), grade_2 = c(2L, 3L, 0L, 11L, 3L, 7L, 2L, 4L,
2L, 1L, 2L, 0L, 1L, 1L, 1L, 0L, 1L, 0L, 2L, 1L, 1L, 0L, 1L, 2L,
1L, 1L, 1L, 0L, NA, NA, NA, NA, NA, NA, NA, NA, NA), grade_3 = c(1L,
4L, 1L, 5L, 3L, 2L, 2L, 5L, 1L, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, 4L, 5L, 1L, NA, NA, NA, 1L, 1L, 0L, 1L, 2L, 0L, 1L, 1L,
0L, 1L, 1L, 0L), grade_4 = c(2L, 4L, 1L, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA)), row.names = c(NA,
37L), class = "data.frame")
CodePudding user response:
Here is a benchmark of the different suggestions if needed :
library(bench)
library(dplyr)
library(data.table)
library(tidyr)
df <- data.frame(
adverse_event = c(
"Haemorrhage", "related", "likely related",
"Other", "related", "likely related", "Pain", "related", "likely related",
"Subcapsular hematoma", "related", "likely related", "Ascites",
"related", "likely related", "Hyperbilirubinemia", "related",
"likely related", "Liver abscess", "related", "likely related",
"Pleural effusion with drainage", "related", "likely related",
"Pneumothorax", "related", "likely related", "Biliary leakage / occlusion / fistula",
"related", "likely related", "Portal vein thrombosis", "related",
"likely related", "Sepsis", "related", "likely related"
),
grade_1 = c(
4L, 4L, 0L, 3L, 6L, 1L, 8L, 4L, 5L, 3L, 1L, 3L, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA
),
grade_2 = c(
2L, 3L, 0L, 11L, 3L, 7L, 2L, 4L, 2L, 1L, 2L, 0L, 1L, 1L, 0L,
1L, 0L, 2L, 1L, 1L, 0L, 1L, 2L, 1L, 1L, 1L, 0L, NA, NA, NA, NA,
NA, NA, NA, NA, NA
),
grade_3 = c(
1L, 4L, 1L, 5L, 3L, 2L, 2L, 5L, 1L, NA, NA, NA, NA, NA, NA,
NA, NA, NA, 4L, 5L, 1L, NA, NA, NA, 1L, 1L, 0L, 1L, 2L, 0L, 1L,
1L, 0L, 1L, 1L, 0L
),
grade_4 = c(
2L, 4L, 1L, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA
)
)
paul_carteron <- function(df){
# where start each group
id <- grep('related', df$adverse_event, invert = T)
# size of each group
size <- lead(id) - id
size_of_last_group <- nrow(df) - id[length(id)] 1
size[length(size)] <- size_of_last_group
# add col with id
df$id <- paste0(rep(df$adverse_event[id], times = size),
df$adverse_event)
# order
df <- df[order(df$id), ]
# remove id
df$id <- NULL
}
lang_tang_dplyr <- function(df){
left_join(
df,
df %>%
filter(!grepl('related', adverse_event)) %>%
select(adverse_event) %>%
arrange(adverse_event) %>%
mutate(o = row_number())
) %>%
mutate(o = data.table::nafill(o, "locf")) %>%
arrange(o) %>%
select(-o)
}
lang_tang_databable <- function(df) {
setDT(df)
data.table(adverse_event = sort(df[!grepl('related',adverse_event), adverse_event]))[, o:=.I][
df, on="adverse_event"][, o:=nafill(o, "locf")][order(o), !c("o")]
}
andre_wilberg <- function(df){
df %>%
mutate(ord = !grepl("related", adverse_event),
grp = cumsum(ord),
Rank = rank(adverse_event[ord])[grp]) %>%
arrange(Rank) %>%
select(-c(ord, grp, Rank))
}
thotal <- function(df){
df %>%
mutate(grp = if_else(grepl("related", adverse_event),
NA_character_,
adverse_event)) %>%
fill(grp) %>%
nest(data = -grp) %>%
arrange(grp) %>%
unnest(cols = data) %>%
select(-grp)
}
results = bench::mark(
iterations = 1000, check = FALSE, time_unit = "s", filter_gc = FALSE,
paul_carteron = paul_carteron(df),
lang_tang_dplyr = lang_tang_dplyr(df),
lang_tang_databable = lang_tang_databable(df),
andre_wilberg = andre_wilberg(df),
thotal = thotal(df)
)
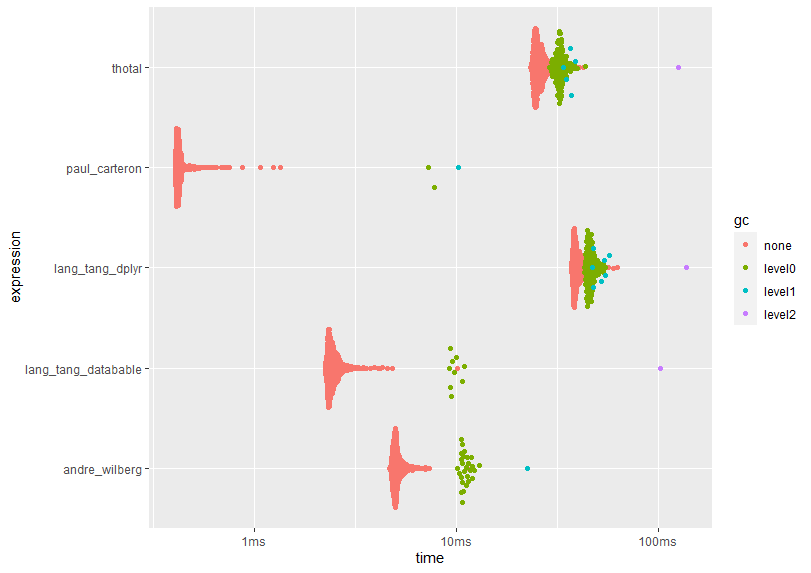
plot(results)
CodePudding user response:
Add your "group" and sort
df$grp=cumsum(!grepl("related",df$adverse_event))
df[order(df$grp,df$adverse_event),]
adverse_event grade_1 grade_2 grade_3 grade_4 grp
1 Haemorrhage 4 2 1 2 1
3 likely related 0 0 1 1 1
2 related 4 3 4 4 1
6 likely related 1 7 2 NA 2
4 Other 3 11 5 NA 2
5 related 6 3 3 NA 2
9 likely related 5 2 1 NA 3
7 Pain 8 2 2 NA 3
8 related 4 4 5 NA 3
12 likely related 3 0 NA NA 4
11 related 1 2 NA NA 4
10 Subcapsular hematoma 3 1 NA NA 4
13 Ascites NA 1 NA NA 5
15 likely related NA 0 NA NA 5
14 related NA 1 NA NA 5
16 Hyperbilirubinemia NA 1 NA NA 6
18 likely related NA 2 NA NA 6
17 related NA 0 NA NA 6
21 likely related NA 0 1 NA 7
19 Liver abscess NA 1 4 NA 7
20 related NA 1 5 NA 7
24 likely related NA 1 NA NA 8
22 Pleural effusion with drainage NA 1 NA NA 8
23 related NA 2 NA NA 8
27 likely related NA 0 0 NA 9
25 Pneumothorax NA 1 1 NA 9
26 related NA 1 1 NA 9
28 Biliary leakage / occlusion / fistula NA NA 1 NA 10
30 likely related NA NA 0 NA 10
29 related NA NA 2 NA 10
33 likely related NA NA 0 NA 11
31 Portal vein thrombosis NA NA 1 NA 11
32 related NA NA 1 NA 11
36 likely related NA NA 0 NA 12
35 related NA NA 1 NA 12
34 Sepsis NA NA 1 NA 12