The documentation for the dplyr package for mutating joins states that for using "by" that "A character vector of variables to join by" is needed.
This however does not seem to be the case.
It seems to work with a numeric or a dbl variable, i.e. column. By this I mean the common column (idenifier) that is used to join the dataframes.
This a glimpse of a portion of the two dataframes. nomem_encr is the common identifier.
df1
Rows: 2,525
Columns: 11
$ nomem_encr <dbl> 800054, 800170, 800186, 800204, 800228, 800274,
$ mj16a093 <dbl lbl> 4, 4, 3, 4, 2, 5, 5, 3, 5, 3, 6,
$ mj16a094 <dbl lbl> 3, 4, 2, 4, 2, 5, 5, 2, 3, 2, 6,
df2
Rows: 6,092
Columns: 3
$ nomem_encr <dbl> 800009, 800015, 800042, 800054, 800057, 800085,
$ cv16h101 <dbl lbl> 2, 3, 5, 6, 7, 6, 0, 6, 5,
$ cv16h044 <dbl lbl> 6, 7, 7, 8, 0, 7, 5, 4, 7, 8, 7,
df3 <- left_join(df1, df2, by = "nomem_encr")
Is it best to convert to a character, or does it not matter? My assumption was that the values just needed to be unique identifiers.
CodePudding user response:
"by" : "A character vector of variables to join by"`
This means that when you write the join, your by argument needs to be a character vector and NOT your column to be a character. for example
left_join(d1, d2, by = c('ID', 'Latitude'))
You are passing a character vector to the by argument. The columns being numeric does not matter
CodePudding user response:
Using Left Join
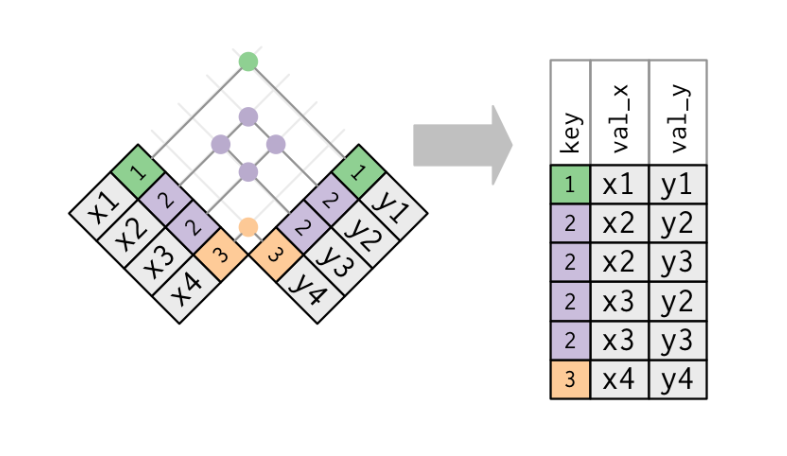
The wording could probably be better, but what is meant by the help page is that you have to supply a character vector for the name of the variable, but the variable type can be anything. Basically you are telling the by argument "join by this variable explicitly". A visual from R for Data Science by Hadley Wickham may help illustrate how this is accomplished with left_join, where the "key" is the by column necessary for joining:
An easy example can be found with the nycflights13 datasets. First you can load the tidyverse and nycflights13 packages.
#### Load Library ####
library(tidyverse)
library(nycflights13)
Then you can check which columns are shared by the flights dataset and the planes dataset.
#### Check Joinable Columns ####
colnames(flights)
colnames(planes) # share tail number
We find they both contain "tailnum":
> colnames(flights)
[1] "year" "month" "day" "dep_time"
[5] "sched_dep_time" "dep_delay" "arr_time" "sched_arr_time"
[9] "arr_delay" "carrier" "flight" "tailnum"
[13] "origin" "dest" "air_time" "distance"
[17] "hour" "minute" "time_hour"
> colnames(planes) # share tail number
[1] "tailnum" "year" "type" "manufacturer" "model"
[6] "engines" "seats" "speed" "engine"
We can combine both datasets with by via a left_join with the tailnum variable like so:
#### Join Two Dataframes ####
flight2 <- flights %>%
left_join(planes,
by = "tailnum")
flight2
Which gives us our combined data:
# A tibble: 336,776 × 27
year.x month day dep_time sched_de…¹ dep_d…² arr_t…³ sched…⁴ arr_d…⁵ carrier
<int> <int> <int> <int> <int> <dbl> <int> <int> <dbl> <chr>
1 2013 1 1 517 515 2 830 819 11 UA
2 2013 1 1 533 529 4 850 830 20 UA
3 2013 1 1 542 540 2 923 850 33 AA
4 2013 1 1 544 545 -1 1004 1022 -18 B6
5 2013 1 1 554 600 -6 812 837 -25 DL
6 2013 1 1 554 558 -4 740 728 12 UA
7 2013 1 1 555 600 -5 913 854 19 B6
8 2013 1 1 557 600 -3 709 723 -14 EV
9 2013 1 1 557 600 -3 838 846 -8 B6
10 2013 1 1 558 600 -2 753 745 8 AA
# … with 336,766 more rows, 17 more variables: flight <int>, tailnum <chr>,
# origin <chr>, dest <chr>, air_time <dbl>, distance <dbl>, hour <dbl>,
# minute <dbl>, time_hour <dttm>, year.y <int>, type <chr>,
# manufacturer <chr>, model <chr>, engines <int>, seats <int>, speed <int>,
# engine <chr>, and abbreviated variable names ¹sched_dep_time, ²dep_delay,
# ³arr_time, ⁴sched_arr_time, ⁵arr_delay
Generalizability of Other Joins
This is accomplished by all other joins in dplyr by the way, but how it does this is different depending on the join you use. The left_join function includes all rows in x (the first dataframe used, in this case flights) whereas a right_join joins by y (the second dataframe planes). Here are the three other examples, which I have labeled by their usage:
#### Alternative Joins ####
flights %>%
right_join(planes,
by = "tailnum") # includes all rows in y
flights %>%
inner_join(planes,
by = "tailnum") # includes all rows in x AND y
flights %>%
full_join(planes,
by = "tailnum") # includes all rows in x OR y
Play around with them and see which is the best for your use case.