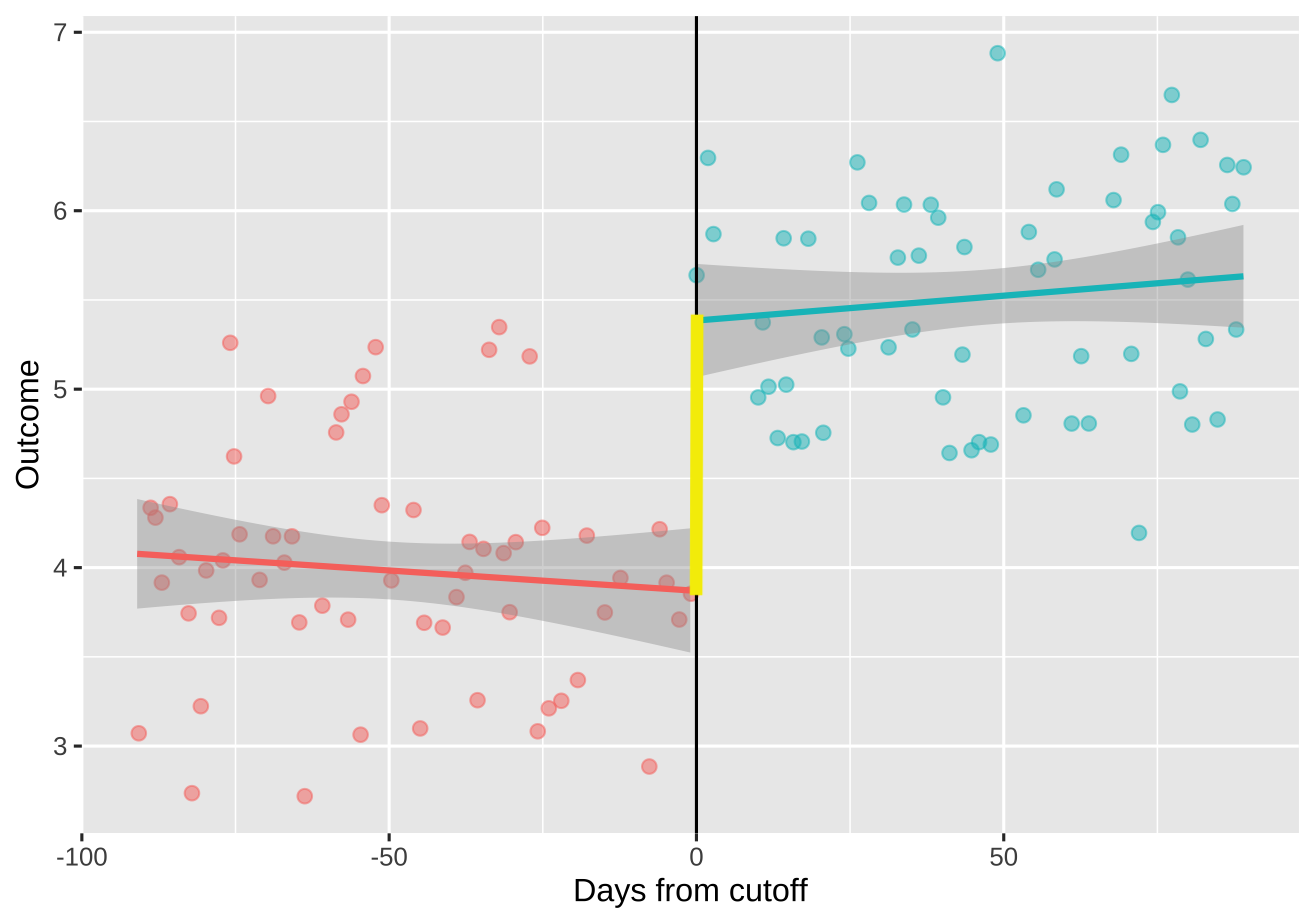
I want to add a vertical line at days==0 that extends from the value of y where days==0 among x==0, to the value y where days==0 among x==1.
df <- structure(list(y = c(3, 4, 4, 3, 4, 4, 4, 3, 4, 4, 4, 3, 4, 4,
4, 3, 3, 4, 3, 3, 4, 4, 4, 3, 4, 3, 3, 4, 4, 3, 4, 5, 4, 4, 4,
5, 4, 5, 4, 4, 5, 4, 4, 5, 4, 4, 5, 4, 4, 4, 5, 5, 4, 4, 4, 4,
5, 5, 5, 4, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5,
5, 4, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 6, 6, 6, 7, 6, 6, 6, 7,
6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6,
6), x = structure(c(1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L), .Label = c("0",
"1"), class = "factor"), days = c(-8, -50, -84, -91, -69, -87,
-89, -19, -61, -18, -46, -26, -35, -51, -88, -55, -36, -44, -24,
-45, -78, -41, -38, -81, -74, -22, -82, -86, -39, -64, -66, -58,
-25, -5, -29, -34, -30, -75, -57, -37, -32, -77, -31, -59, -67,
-83, -70, -1, -65, -15, -27, -56, -71, -80, -12, -3, -76, -54,
-52, -6, 35, 20, 53, 61, 43, 71, 88, 31, 17, 85, 21, 25, 16,
46, 45, 41, 15, 48, 72, 63, 24, 12, 83, 40, 13, 10, 11, 79, 81,
64, 38, 59, 3, 77, 39, 26, 68, 49, 87, 69, 75, 33, 34, 76, 78,
86, 14, 36, 0, 44, 54, 58, 18, 80, 82, 89, 56, 2, 28, 74)), row.names = c(NA,
-120L), class = c("tbl_df", "tbl", "data.frame"))
# https://evalf20.classes.andrewheiss.com/example/rdd/
library(tidyverse)
df %>%
ggplot(aes(x = days, y = y, color = x))
geom_point(size = 2, alpha = 0.5, position = position_jitter(seed = 42))
geom_smooth(data = filter(df, days < 0), method = "lm")
geom_smooth(data = filter(df, days >= 0), method = "lm")
geom_vline(xintercept = 0)
labs(x = "Days from cutoff", y = "Outcome")
guides(color = FALSE)
The slopes of each line can vary, so we can't assume for instance that the value of y at days==0 among x==0 (left) is always at the min of y for that segment as shown.
CodePudding user response:
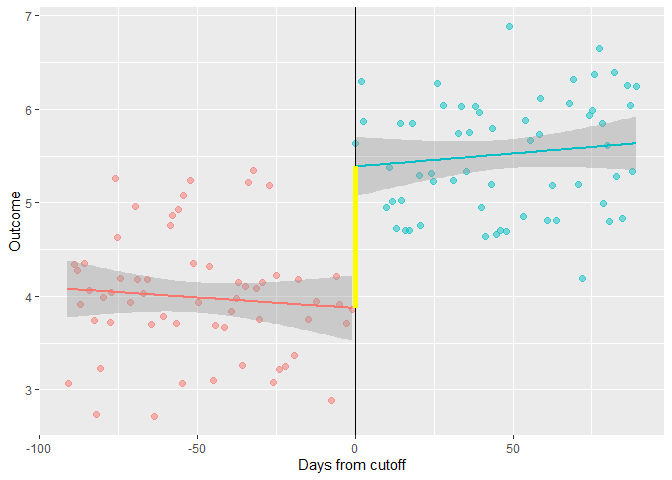
You can extract the intercept for the geom_smooth regression lines and add them to your plot as a geom_segment:
library(tidyverse)
df %>%
ggplot(aes(x = days, y = y, color = x))
geom_point(size = 2, alpha = 0.5, position = position_jitter(seed = 42))
geom_smooth(data = filter(df, days < 0), method = "lm")
geom_smooth(data = filter(df, days >= 0), method = "lm")
geom_vline(xintercept = 0)
labs(x = "Days from cutoff", y = "Outcome")
guides(color = "none") -> gg
lm(y~days, data = filter(df, days < 0))-> lm_neg
lm(y~days, data = filter(df, days >= 0))-> lm_pos
gg
geom_segment(aes(x = 0, y = lm_neg$coefficients[1],
xend = 0, yend = lm_pos$coefficients[1]),
colour = "yellow", size = 2)
CodePudding user response:
df <- structure(list(y = c(3, 4, 4, 3, 4, 4, 4, 3, 4, 4, 4, 3, 4, 4,
4, 3, 3, 4, 3, 3, 4, 4, 4, 3, 4, 3, 3, 4, 4, 3, 4, 5, 4, 4, 4,
5, 4, 5, 4, 4, 5, 4, 4, 5, 4, 4, 5, 4, 4, 4, 5, 5, 4, 4, 4, 4,
5, 5, 5, 4, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5,
5, 4, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 6, 6, 6, 7, 6, 6, 6, 7,
6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6,
6), x = structure(c(1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L), .Label = c("0",
"1"), class = "factor"), days = c(-8, -50, -84, -91, -69, -87,
-89, -19, -61, -18, -46, -26, -35, -51, -88, -55, -36, -44, -24,
-45, -78, -41, -38, -81, -74, -22, -82, -86, -39, -64, -66, -58,
-25, -5, -29, -34, -30, -75, -57, -37, -32, -77, -31, -59, -67,
-83, -70, -1, -65, -15, -27, -56, -71, -80, -12, -3, -76, -54,
-52, -6, 35, 20, 53, 61, 43, 71, 88, 31, 17, 85, 21, 25, 16,
46, 45, 41, 15, 48, 72, 63, 24, 12, 83, 40, 13, 10, 11, 79, 81,
64, 38, 59, 3, 77, 39, 26, 68, 49, 87, 69, 75, 33, 34, 76, 78,
86, 14, 36, 0, 44, 54, 58, 18, 80, 82, 89, 56, 2, 28, 74)), row.names = c(NA,
-120L), class = c("tbl_df", "tbl", "data.frame"))
# https://evalf20.classes.andrewheiss.com/example/rdd/
library(tidyverse)
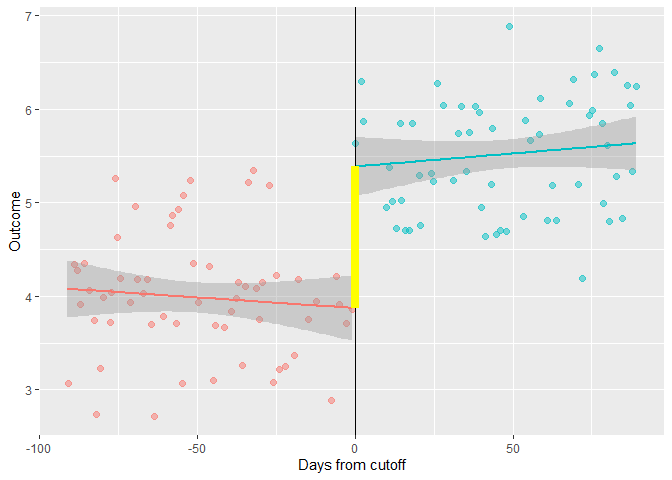
y1 = predict(lm(y ~ days, filter(df, days < 0)), list(days=0))
y2 = predict(lm(y ~ days, filter(df, days >= 0)), list(days=0))
df %>%
ggplot(aes(x = days, y = y, color = x))
geom_point(size = 2, alpha = 0.5, position = position_jitter(seed = 42))
geom_smooth(data = filter(df, days < 0), method = "lm")
geom_smooth(data = filter(df, days >= 0), method = "lm")
geom_vline(xintercept = 0)
labs(x = "Days from cutoff", y = "Outcome")
guides(color = FALSE)
annotate("segment", x=0,xend=0, y=y1, yend=y2 , color = "yellow", size = 3)
Created on 2023-01-16 with reprex v2.0.2
