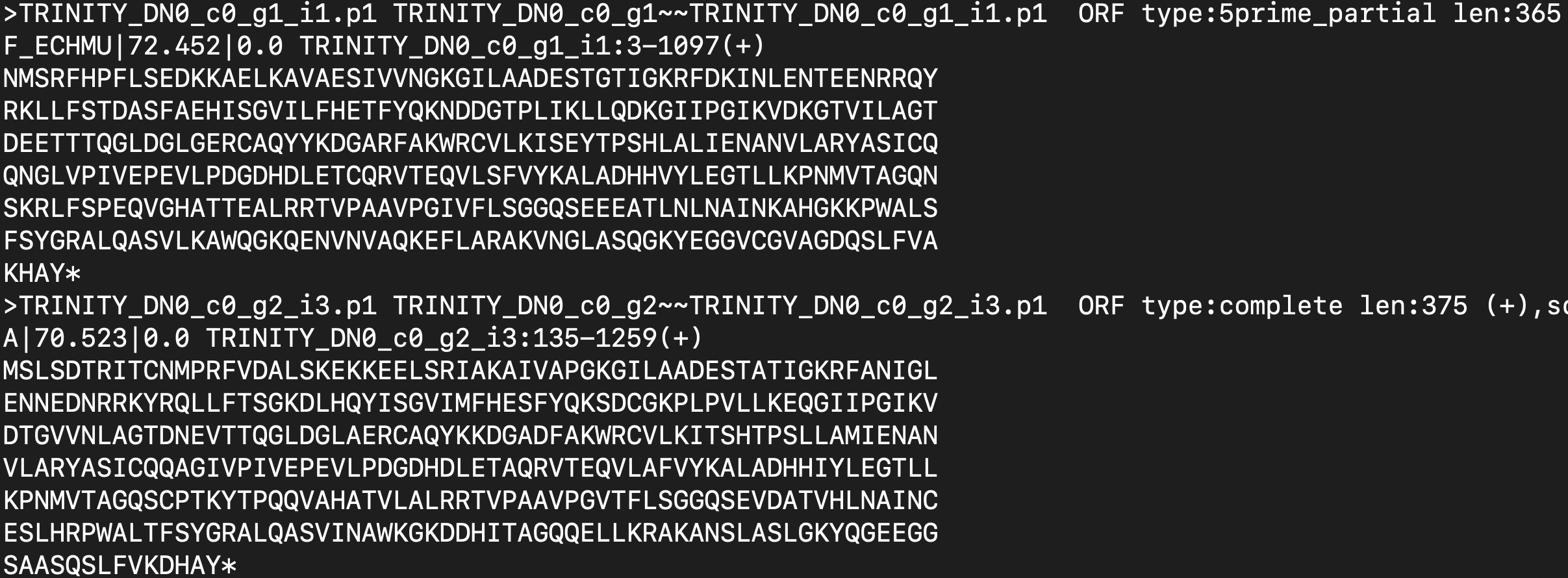
I'm trying to extract just the sequence ID from the the file in Linux server.
To give you few examples TRINITY_DN0_c0_g1_i1.p1 and TRINITY_DN0_c0_g1_i3.p1 and sequence IDs. The sequence ID lengths are not same but they all start with TRINITY and ends with .p1.
I tried using awk '{print$1}' filename.cdhit > seq_id.fasta but instead I got this

I just want the ID but it would also extract non-interest information as well (the lengthy alphabetic protein seq). I have attempted to create a python script in hopes to just extract the IDs:
import re
file_path = '/var2/user/de_novo/data/transdecoder_dir/Trinity.fasta.transdecoder.pep.cdhi
t'
new_file_path = '/var2/user/de_novo/data/transdecoder_dir/seqID.fasta'
with open(file_path, 'rt') as file:
for myline in file:
if "\.p1" in file:
with open(new_file_path, 'w') as new_file:
new_file.write()
else:
print('No match found.')
Tried creating python script, running linux command But comes out as not match found. Not sure where I went wrong. Would appreciate with any help, thank you.
CodePudding user response:
This should do the trick:
awk '{print$1}' filename.cdhit | grep TRINITY | cut -c2-
As for python, check out https://biopython.org/. It's a collection of bioinformatics-related goodies.
OR
Here a '.cdhit'-file reader: https://pypi.org/project/cdhit-reader/