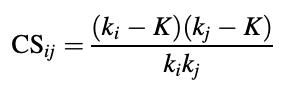I have this great presence-absence dataset in which I need to calculate a C score (CS) for each species pair (BABO, BW, RC, SKS, MANG) to measure species co-occurrences.
ki and kj denote the numbers of occurrences of species i and j and K is the number of co-occurrences of both species.
I have looked at articles like Find the pair of most correlated variables and Turning co-occurrence counts into co-occurrence probabilities with cascalog but was unable to determine the most efficient way to go about it in R. I have tried creating a function but was unsuccessful.
My data:
data <- structure(list(group_id = c("2008-2-11.C3_900", "2008-2-11.C3_960",
"2008-2-11.C3_1200", "2008-2-11.C3_1230", "2008-2-11.C3_1460",
"2008-2-11.C3_1490", "2008-2-22.Mwani_0", "2008-2-22.Mwani_110",
"2008-2-22.Mwani_600", "2008-2-22.Mwani_1650", "2008-2-20.Sanje_150",
"2008-2-20.Sanje_410", "2008-2-20.Sanje_3000", "2008-5-9.C3_900",
"2008-5-13.Mwani_750", "2008-5-13.Mwani_800", "2008-5-13.Mwani_900",
"2008-5-13.Mwani_1080", "2008-5-13.Mwani_1800", "2008-5-13.Mwani_2200",
"2008-5-13.Mwani_2900"), BABO = c(0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0), BW = c(1, 1, 1, 1, 0, 0,
0, 0, 1, 0, 1, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1), RC = c(0, 0, 1,
1, 1, 0, 0, 1, 0, 0, 0, 1, 1, 0, 0, 1, 1, 1, 0, 0, 0), SKS = c(0,
1, 1, 0, 0, 1, 1, 0, 0, 1, 1, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0),
MANG = c(0, 0, 0, 0, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 1)), row.names = c(NA, -21L), class = c("tbl_df",
"tbl", "data.frame"))
group_id BABO BW RC SKS MANG
<chr> <dbl><dbl><dbl><dbl>
2008-2-11.C3_900 0 1 0 0 0
2008-2-11.C3_960 0 1 0 1 0
2008-2-11.C3_1200 0 1 1 1 0
2008-2-11.C3_1230 0 1 1 0 0
2008-2-11.C3_1460 0 0 1 0 0
2008-2-11.C3_1490 0 0 0 1 0
2008-2-22.Mwani_0 0 0 0 1 1
2008-2-22.Mwani_110 0 0 1 0 1
2008-2-22.Mwani_600 0 1 0 0 0
2008-2-22.Mwani_1650 0 0 0 1 0
2008-2-20.Sanje_150 0 1 0 1 0
2008-2-20.Sanje_410 0 0 1 0 0
2008-2-20.Sanje_3000 0 0 1 0 0
2008-5-9.C3_900 0 0 0 1 0
2008-5-13.Mwani_750 0 0 0 1 0
2008-5-13.Mwani_800 1 0 1 0 0
2008-5-13.Mwani_900 0 0 1 0 0
2008-5-13.Mwani_1080 0 1 1 0 0
2008-5-13.Mwani_1800 0 1 0 0 0
2008-5-13.Mwani_2200 0 1 0 0 0
2008-5-13.Mwani_2900 0 1 0 0 1
CodePudding user response:
First define a function to compute CS for a given pair of species; then use combn() to generate all possible pairs; then pass each pair to the CS() function using purrr::map2_dbl().
library(dplyr)
library(purrr)
CS <- function(i, j, data) {
K <- sum(data[[i]] == 1 & data[[j]] == 1)
ki <- sum(data[[i]])
kj <- sum(data[[j]])
((ki - K) * (kj - K)) / (ki * kj)
}
names(sp_data)[-1] %>%
combn(2) %>%
t() %>%
as_tibble() %>%
set_names(c("i", "j")) %>%
mutate(CS = map2_dbl(i, j, CS, data = sp_data))
# A tibble: 10 × 3
i j CS
<chr> <chr> <dbl>
1 BABO BW 1
2 BABO RC 0
3 BABO SKS 1
4 BABO MANG 1
5 BW RC 0.467
6 BW SKS 0.438
7 BW MANG 0.6
8 RC SKS 0.778
9 RC MANG 0.593
10 SKS MANG 0.583
(Side note: using the equation you provided, species with little co-occurrence end up with high CS scores, and vice versa. If this isn’t what you expected, perhaps your equation should be subtracted from 1?)
CodePudding user response:
In base R, we may use crossprod
t1 <- crossprod(as.matrix(data[-1]))
v1 <- diag(t1)
c1 <- v1[col(t1)]
r1 <- v1[row(t1)]
((c1 - t1) * (r1 - t1))/(c1 * r1)
-output
BABO BW RC SKS MANG
BABO 0 1.0000000 0.0000000 1.0000000 1.0000000
BW 1 0.0000000 0.4666667 0.4375000 0.6000000
RC 0 0.4666667 0.0000000 0.7777778 0.5925926
SKS 1 0.4375000 0.7777778 0.0000000 0.5833333
MANG 1 0.6000000 0.5925926 0.5833333 0.0000000