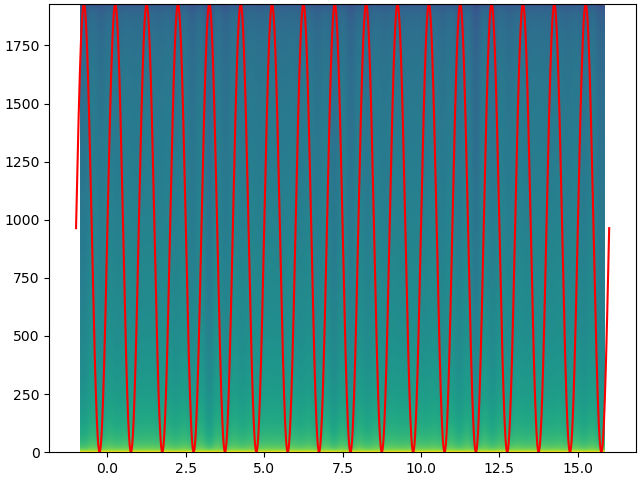
I have signal which starts at time -1 seconds.
However, when plotting the spectrogram, the first bin edge starts at 0 (midpoint at 0.25) How do I change this so that my data is represented accurately when plotting on the x-axis?
Using xextent=(time[0] 0.125, time[-1]) seems to fix this problem. However, I am unsure what variable determines the bin width, and therefore concerned this might change with other datasets of different sampling rate, number of points, etc.
Example code:
import numpy as np
import matplotlib.pyplot as plt
import signal
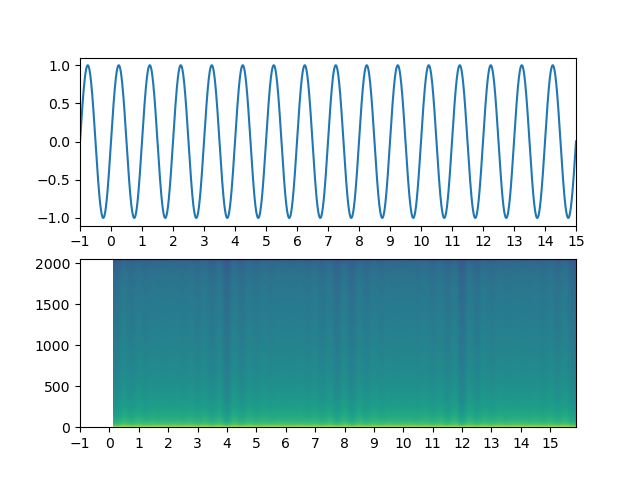
time = np.linspace(-1, 16, 65536)
signal = np.sin(2 * np.pi * time)
fig = plt.figure()
ax_top = fig.add_subplot(211)
ax_spec = fig.add_subplot(212)
ax_top.plot(time, signal)
ax_top.set_xlim(-1, 10)
ax_spec.set_xlim(-1, 10)
Pxx, freqs, bins, cax = ax_spec.specgram(signal, NFFT=2048, Fs=4096, noverlap=2048 / 2, mode='magnitude', pad_to=2048 * 16)
ax_spec.set_xticks(np.arange(time[0], time[-1], 1))
ax_top.set_xticks(np.arange(time[0], time[-1], 1))
plt.show()
CodePudding user response:
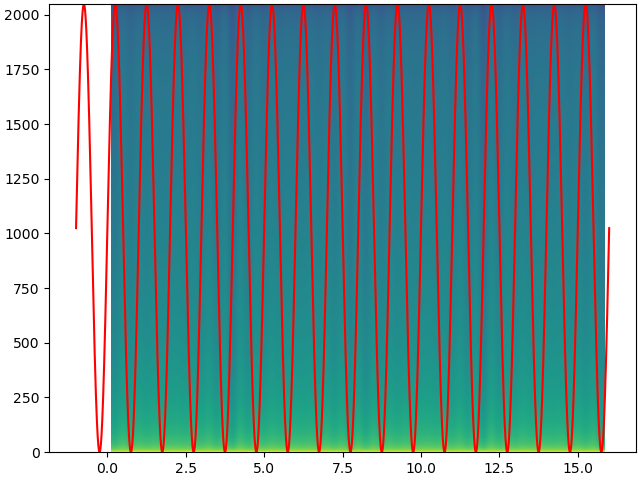
I will simplify your example slightly, with less manual settings to make sure the axes come out correctly out of the box:
import numpy as np
import matplotlib.pyplot as plt
time = np.linspace(-1, 16, 65536)
signal = np.sin(2 * np.pi * time)
fig, ax = plt.subplots(constrained_layout=True)
# In your case, use this instead:
#fig, (ax_top, ax_spec) = plt.subplots(2, 1, constrained_layout=True)
ax.plot(time, 1024 * signal 1024, c='r')
Pxx, freqs, bins, cax = ax.specgram(signal, NFFT=2048, Fs=4096, noverlap=2048/2, mode='magnitude', pad_to=2048*16)
plt.show()
The goal is to get the time axes of the signal and the image co-aligned.
The important part of the spectrogram code is implemented in