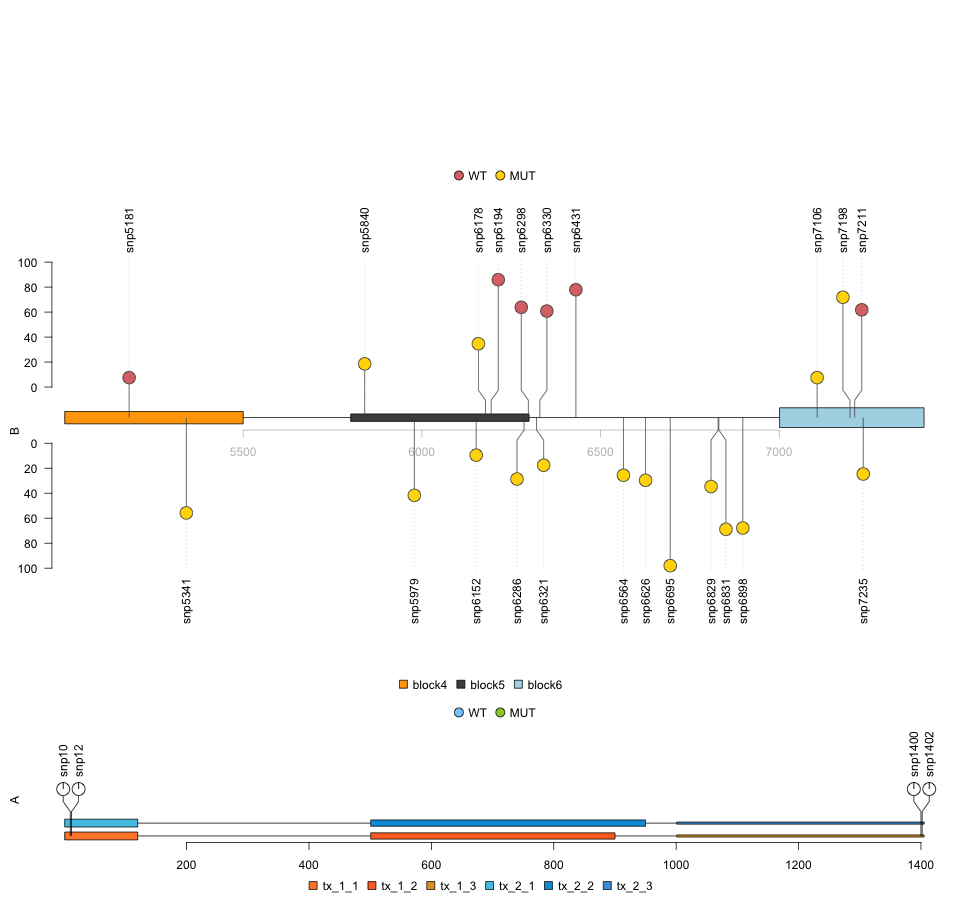
I'm trying to move the Lolliplot so that all of the labels are on the plot.
I am following the vignette here 
CodePudding user response:
If you specify the size of the png you can ensure the plot 'fits', e.g. (from the vignette):
library(Gviz)
library(rtracklayer)
library(trackViewer)
extdata <- system.file("extdata", package="trackViewer",
mustWork=TRUE)
SNP <- c(10, 12, 1400, 1402)
sample.gr <- GRanges("chr1", IRanges(SNP, width=1, names=paste0("snp", SNP)))
features <- GRanges("chr1", IRanges(c(1, 501, 1001),
width=c(120, 400, 405),
names=paste0("block", 1:3)))
SNP2 <- sample(4000:8000, 30)
x2 <- sample.int(100, length(SNP2), replace=TRUE)
sample2.gr <- GRanges("chr3", IRanges(SNP2, width=1, names=paste0("snp", SNP2)),
value1=x2, value2=100-x2)
sample2.gr$color <- rep(list(c('#DB7575', '#FFD700')), length(SNP2))
sample2.gr$border <- "gray30"
features2 <- GRanges("chr3", IRanges(c(5001, 5801, 7001),
width=c(500, 500, 405),
names=paste0("block", 4:6)),
fill=c("orange", "gray30", "lightblue"),
height=unit(c(0.5, 0.3, 0.8), "cm"))
legends <- list(list(labels=c("WT", "MUT"), fill=c("#87CEFA", '#98CE31')),
list(labels=c("WT", "MUT"), fill=c('#DB7575', '#FFD700')))
rand.id <- sample.int(length(sample.gr), 3*length(sample.gr), replace=TRUE)
rand.id <- sort(rand.id)
sample.gr.mul.patient <- sample.gr[rand.id]
## pie.stack require metadata "stack.factor", and the metadata can not be
## stack.factor.order or stack.factor.first
len.max <- max(table(rand.id))
stack.factors <- paste0("patient", formatC(1:len.max,
width=nchar(as.character(len.max)),
flag="0"))
sample.gr.mul.patient$stack.factor <-
unlist(lapply(table(rand.id), sample, x=stack.factors))
sample.gr.mul.patient$value1 <-
sample.int(100, length(sample.gr.mul.patient), replace=TRUE)
sample.gr.mul.patient$value2 <- 100 - sample.gr.mul.patient$value1
patient.color.set <- as.list(as.data.frame(rbind(rainbow(length(stack.factors)),
"#FFFFFFFF"),
stringsAsFactors=FALSE))
names(patient.color.set) <- stack.factors
sample.gr.mul.patient$color <-
patient.color.set[sample.gr.mul.patient$stack.factor]
legend <- list(labels=stack.factors, col="gray80",
fill=sapply(patient.color.set, `[`, 1))
features.mul <- rep(features, 2)
features.mul$height[4:6] <- list(unit(1/8, "inches"),
unit(0.5, "lines"),
unit(.2, "char"))
features.mul$fill <- c("#FF8833", "#F9712A", "#DFA32D",
"#51C6E6", "#009DDA", "#4B9CDF")
end(features.mul)[5] <- end(features.mul[5]) 50
features.mul$featureLayerID <-
paste("tx", rep(1:2, each=length(features)), sep="_")
names(features.mul) <-
paste(features.mul$featureLayerID,
rep(1:length(features), 2), sep="_")
sample2.gr$score <- sample2.gr$value1
sample2.gr$SNPsideID <- "top"
idx <- sample.int(length(sample2.gr), 15)
sample2.gr$SNPsideID[idx] <- "bottom"
sample2.gr$color[idx] <- '#FFD700'
lolliplot(list(A=sample.gr, B=sample2.gr),
list(x=features.mul, y=features2),
type=c("pie", "circle"), legend=legends)
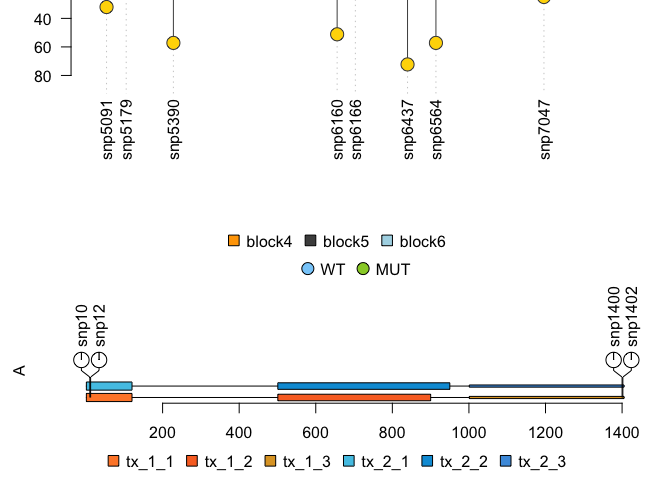
This plot (above) is 'chopped off', but if you save the plot as a png, you can specify the size of the 'canvas' to ensure the plot 'fits' properly:
png("example.png", width = 960, height = 900)
lolliplot(list(A=sample.gr, B=sample2.gr),
list(x=features.mul, y=features2),
type=c("pie", "circle"), legend=legends)
dev.off()
#> quartz_off_screen
#> 2
Created on 2021-11-03 by the reprex package (v2.0.1)